BioTrust Enthusiasts Weekly (Vol. 148):The AI Revolution Shouldn’t Be Compared to the Internet, but to Transistors¶
Here’s a record of weekly raw letter-related content worth sharing, posted on Sundays.
This magazine is open source (GitHub.openbiox/weeklyWe welcome the submission of issues, manuscripts, or letters of recommendation.
“Sangshin Weekly Discussion Forum
cover art¶

Topic of the Week:The AI revolution shouldn’t be compared to the internet, but to transistors¶
The article recaps and carefully compiles an interview with OpenAI CEO Sam Altman, who argues that “AI is incredibly scalable and rapidly penetrating all areas. There’s a lot of transistorized technology behind the products and services you use today, but you wouldn’t think of these companies as ‘transistorized companies’.”
@Wangcy-rachel: Currently, the AI revolution is rapidly embedding itself in our lives and in various industries, and AI will, like transistors, will evolve rapidly and iteratively and be rapidly applied to products in various industries that will greatly facilitate our lives and work.
Sangshin Research¶
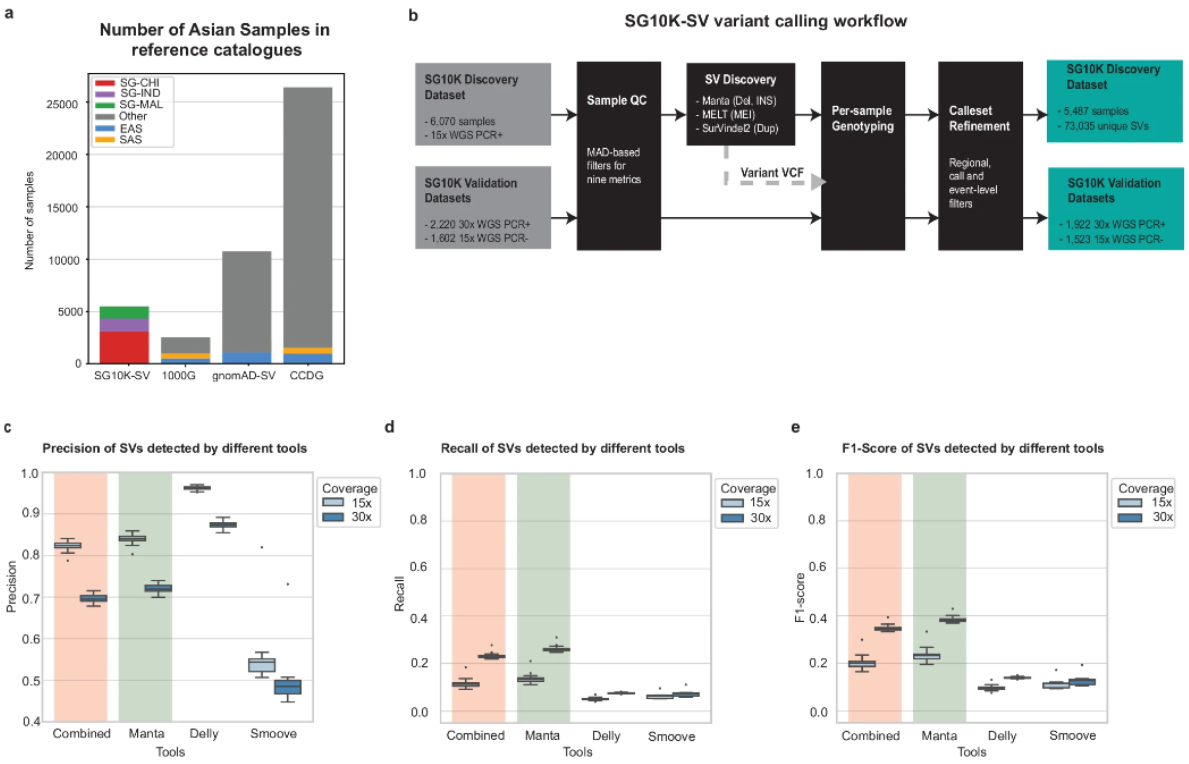
This article is about “Nat Commun Publishes Largest Asian Multi-Ancestry SV Atlas to Date, Revealing Genomic Variability Patterns in Asian Populations”, which introduces a large-scale Asian multi-ancestry SV study published by the Agency for Science, Technology and Research (ASTR), Singapore, and a multi-unit research team in Nature Communications, which fills a critical gap in the genomic variation mapping of Asian populations. This study fills a key gap in the genomic variation mapping of Asian populations, and is of great significance to the understanding of genomic variation patterns in Asian populations.
- 原文链接:https://ift.tt/Vkv4yPW
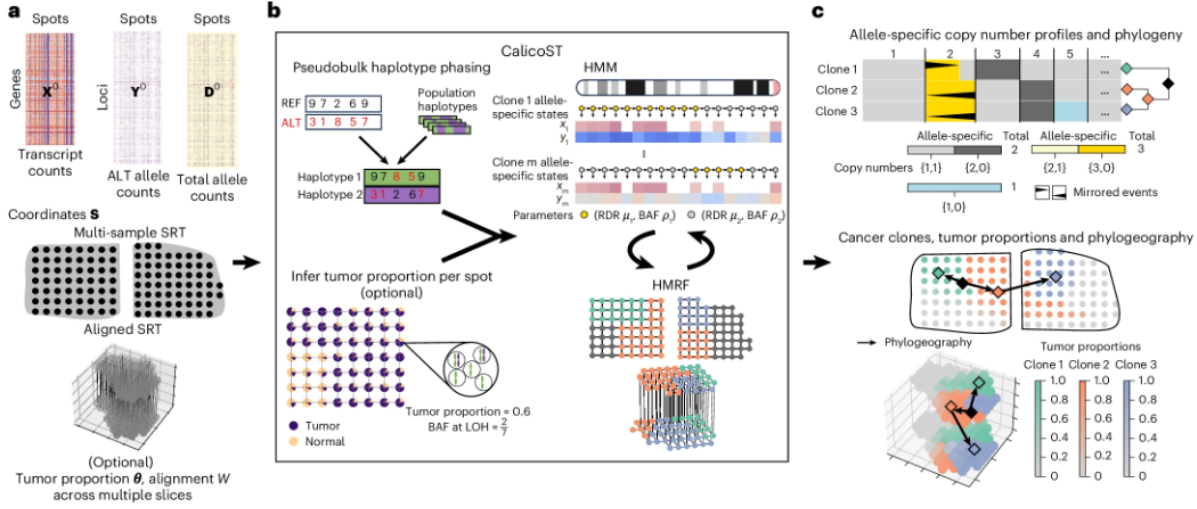
This article is about “Nature Methods publishes new algorithm, CalicoST, to construct ‘spatial evolutionary maps’ of tumors using spatial transcriptome data” Abstract, which focuses on a collaboration between Princeton University and the University of Washington’s A team of researchers has collaborated to develop a new algorithm, CalicoST, which can infer allele-specific copy number variants (CNAs) from spatial transcriptome data and reconstruct the spatial evolutionary trajectory of tumors.
- Original link: https://ift.tt/Hwr7sYN
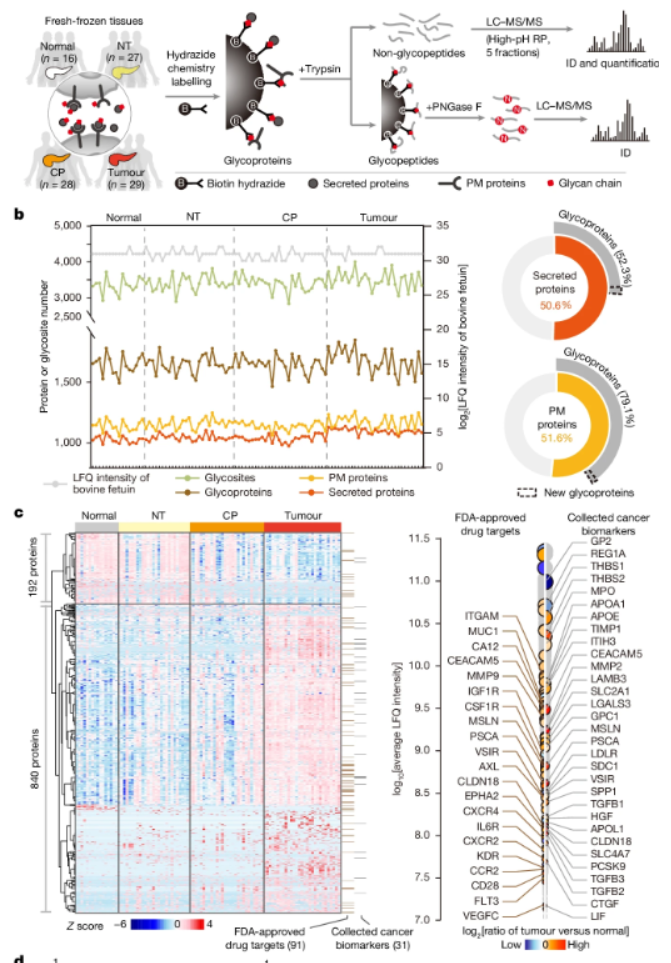
This Nature article develops a multidimensional clinical functional proteomics analysis strategy and applies it to systematically analyze the intercellular signaling network in the tumor microenvironment of pancreatic cancer, providing new ideas and systematic functional proteomic big data for the discovery of new pancreatic cancer markers and drug targets.
- 原文链接:https://ift.tt/1JBE6xP
Bowen Consulting¶
4、A beginner-friendly explanation of the process of analyzing macrogenomic data
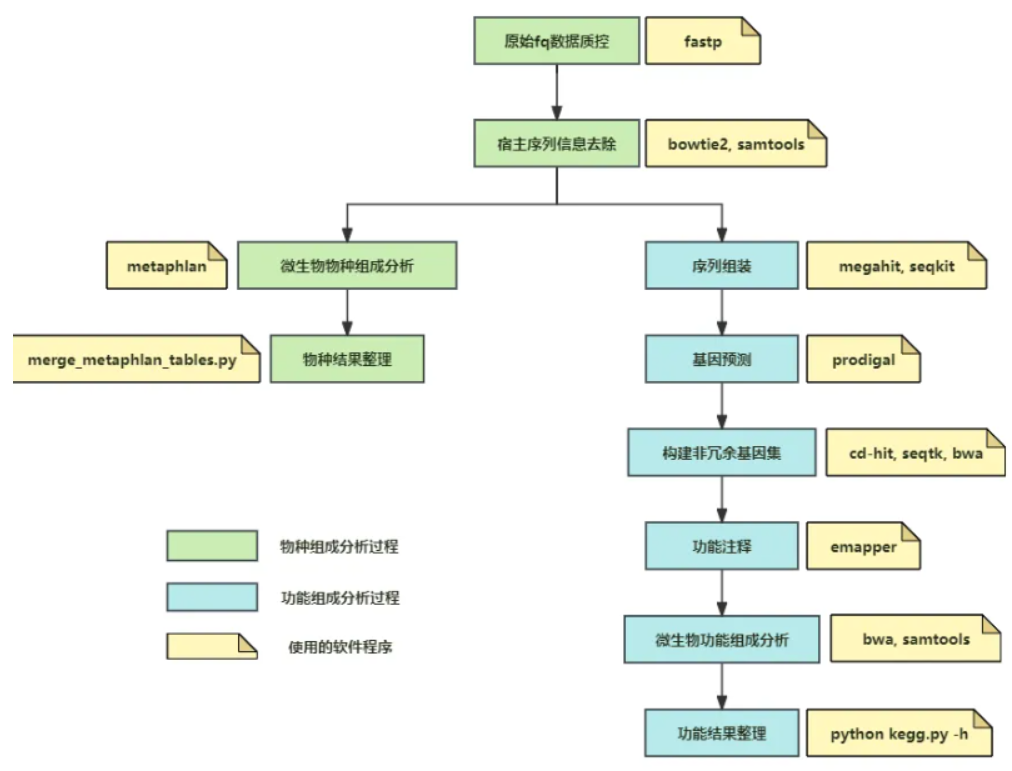
In the field of human intestinal microecology, macro-genome sequencing technology is a highly efficient research tool to rapidly obtain information on the species characteristics of microbiota and the functions they perform. There are many packaged software or packages for macrogenome analysis, but for beginners, directly utilizing these packaged one-stop software or packages is not very friendly to understand the macrogenome analysis process, and there are a lot of “black-box” contents that cannot be understood. This article describes each step of the macro genome analysis process, which is suitable for beginners to understand and learn.
5、B-cell immune checkpoints and cancer therapy
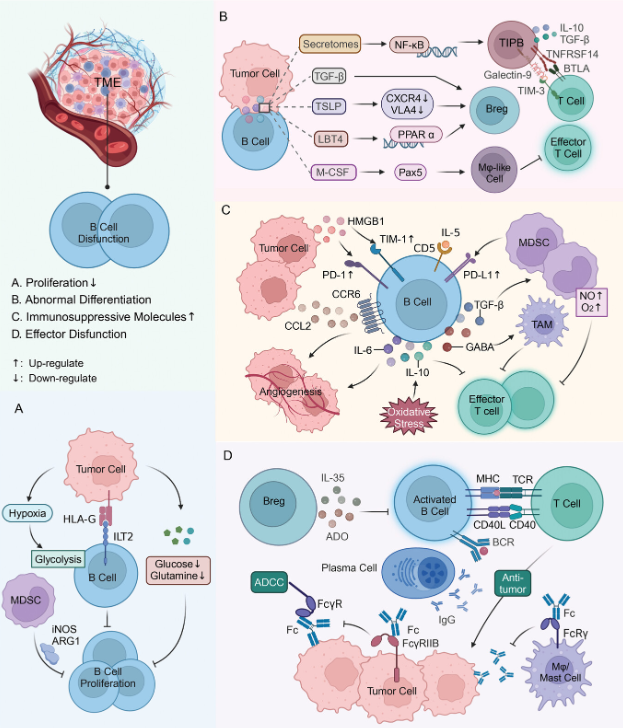
A review on B-cell immune checkpoints and cancer therapy is presented in the article, which can help us to understand the detailed knowledge and cutting-edge hotspots of B-cell immune checkpoints and cancer therapy.
- Original link: https://ift.tt/cLJEI0P
6、Summary of R codes for sample size calculation
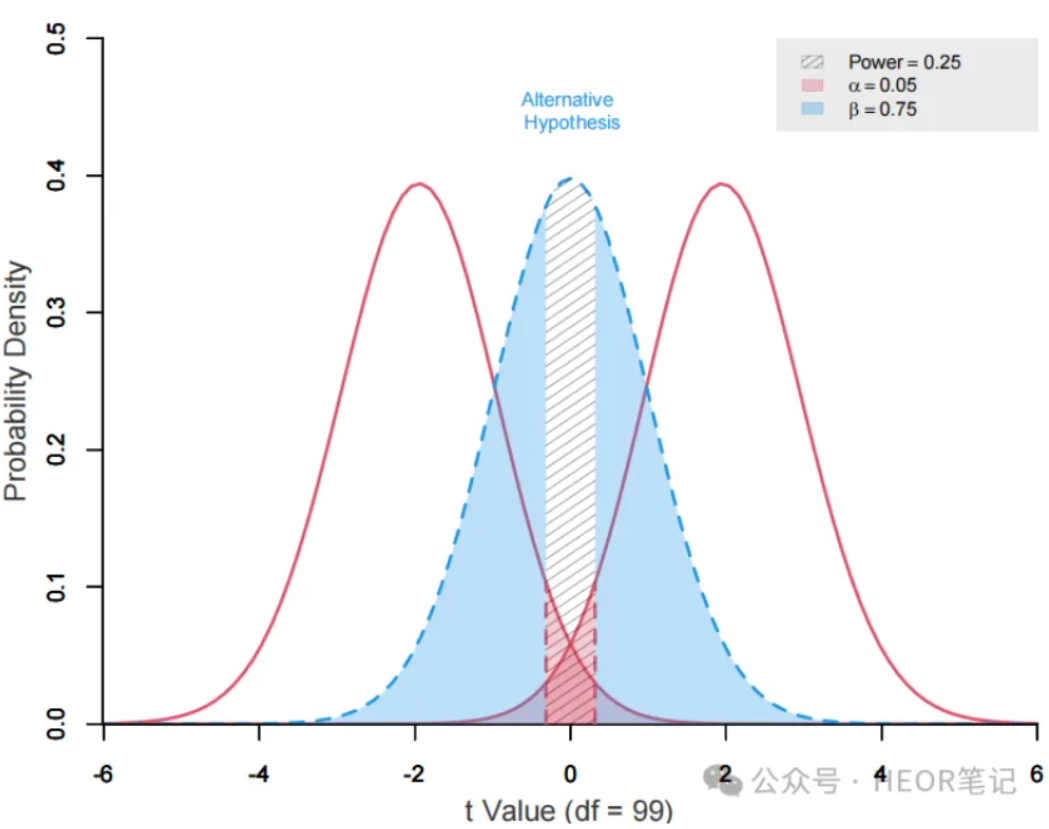
The code for calculating sample sizes for various tests and regressions using the R package pwrss is summarized in detail in the text, along with example plots.
artifact¶
7、Tidyplots | Making Code-Based Data Visualization Easy for Life Scientists
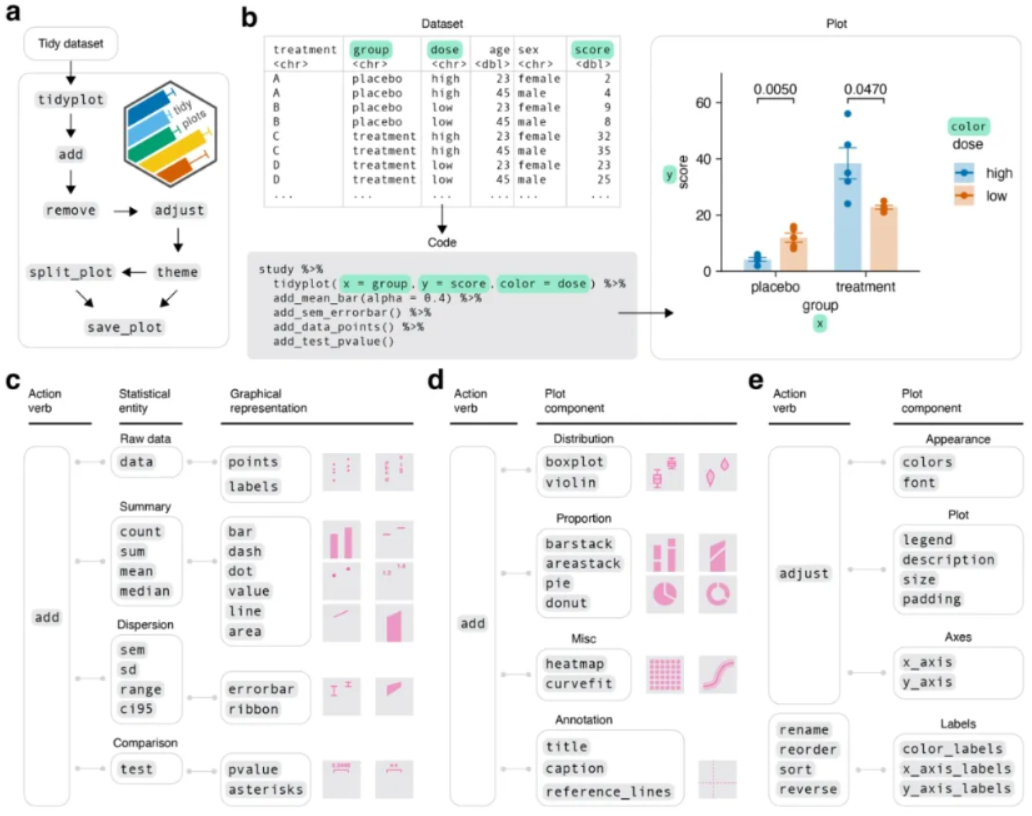
The article describes tidyplots, a user-friendly code-based tool for creating customizable, insightful charts.
- Tools: https://ift.tt/Ghrl70H
8、SplitsTree App | Interactive Analysis and Visualization Using Phylogenetic Trees and Networks
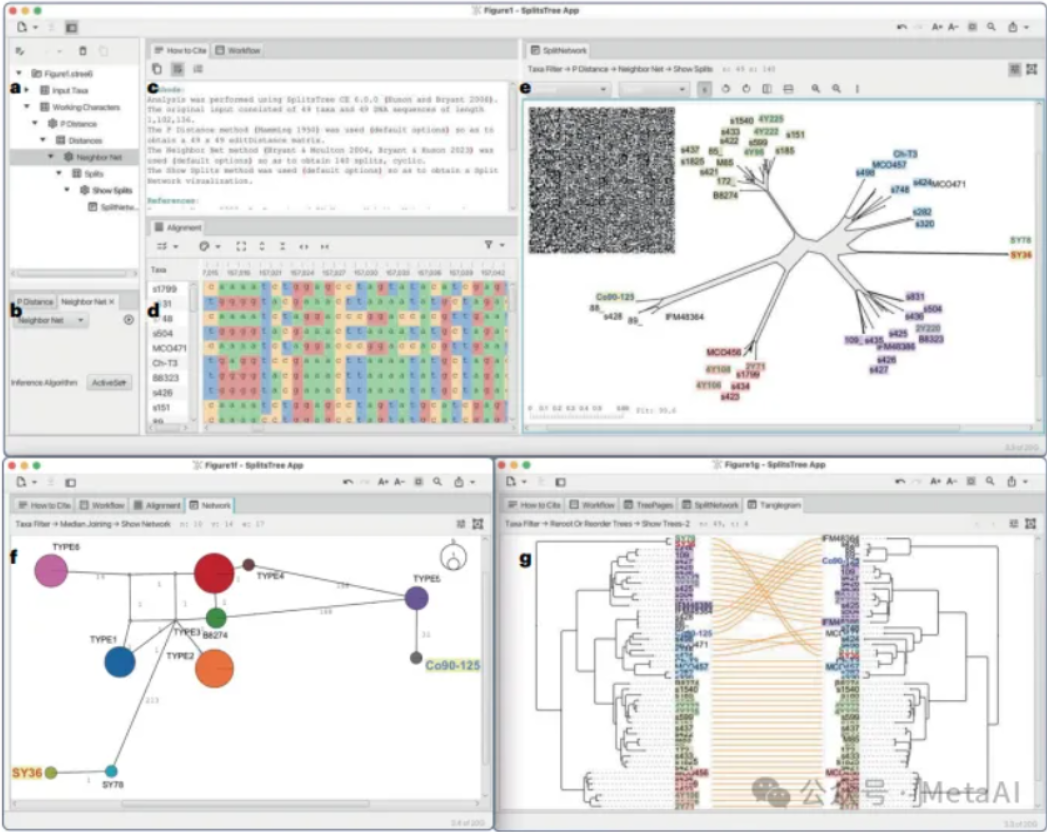
The paper presents SplitsTreeApp, a software for analyzing evolutionary data through phylogenetic trees and networks, integrating more than 100 algorithms and capable of handling a wide range of input formats for different types of data analysis.
- Tools: https://ift.tt/bVUHRsw
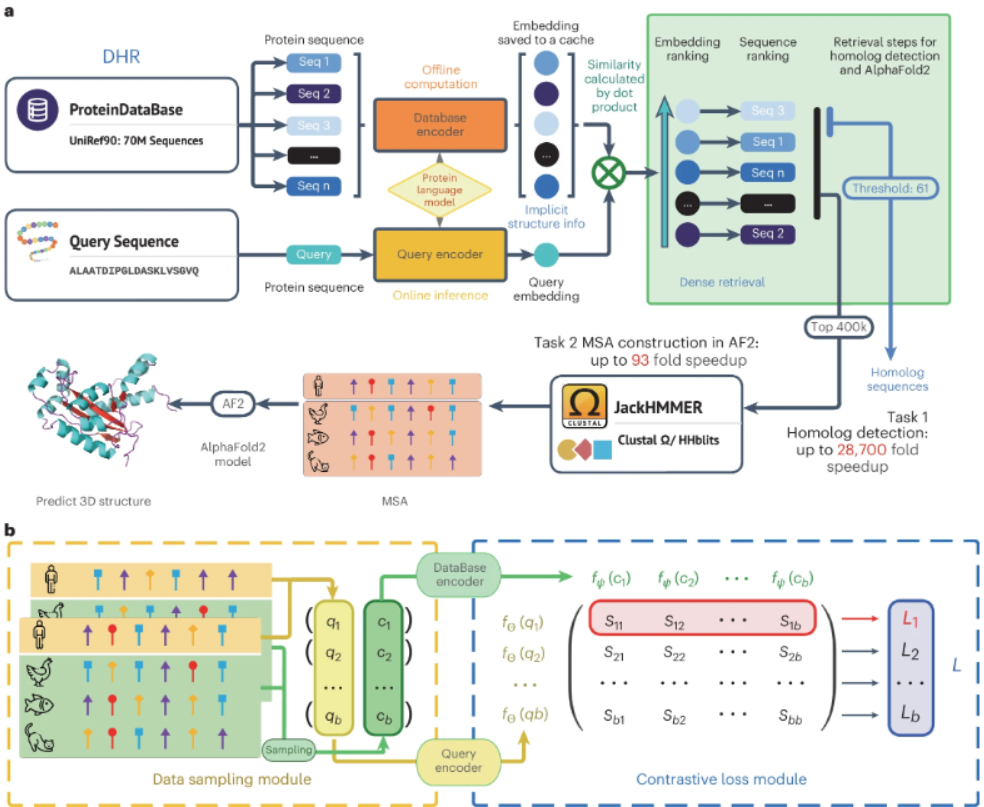
This paper provides an ultra-fast and highly sensitive method, Dense Homolog Retriever (DHR), to detect homologs based on protein language modeling and dense retrieval techniques. Its dual-encoder architecture generates different embeddings for the same protein sequence and easily localizes homologs by pairwise comparison of these representations. Its alignment-free nature improves speed, while the protein language model incorporates rich evolutionary and structural information in DHR embeddings. Compared to previous methods, DHR improves sensitivity by more than 10%, and by more than 56% at the superfamily level for samples that are difficult to recognize using alignment-based methods. It is more than 22-fold faster than conventional methods such as PSI-BLAST and DIAMOND, and more than 28,700-fold faster than HMMER.DHR’s unique discovery of new distant homologs is useful for revealing connections between known proteins and for improving our understanding of protein evolution, structure and function.
- Link to paper: https://ift.tt/5V8rU1G
- 工具:https://ift.tt/BV1XvWM
resource (such as manpower or tourism)¶
10、List of must-read papers on AIGC algorithms

The article contains a detailed summary of must-read papers on different AIGC algorithms, related materials, and a list of links that are worth reading for those in related fields.
可以在 https://ift.tt/4Q3iU5m 下载 PDF。
Contributor (GitHub ID)¶
“Openbiox Weekly” operation and maintenance team:
@ShixiangWang(Wang Shixiang)@kkjtmac(Kan Kogia)@NiEntropy(Zhao Qixiang)@He-Kai-fly(Kay Ho)@JnanZhang(Jannan Zhang)@Tomcxf(Chen Xiaofeng)@wangdepin(Wang Depin)@kongjianyang(Space Yang)@donghongyu2020(Dong Hongyu)@DrRobinLuo(Luo Peng)@Wangcy-rachel– Wang Chunyang@zoe3251– Shu Chenyang (1409-1524), Northern Song dynasty poet
订阅¶
This weekly newsletter is published every Sunday and is synchronized and updated on the WeChat public number “elegant R” (elegant-r).
Search for “Elegant R” on WeChat or scan the QR code to subscribe.
(concluded)