Original link: https://kaopubear.top/blog/2022-12-16-fmi-kars-pancancer-landscape/
Both KRAS and HRAS, as well as NRAS, belong to the RAS family, and these genes, when mutated, can initiate or promote tumor growth. Although KRAS is a common mutated oncogene in many tumors, the protein encoded by the KRAS gene is like a smooth ball, and it is difficult to find small molecular compounds that can catch it. In the past few decades, it has been considered “undruggable”.
However, improvements in drug design methods have allowed scientists to develop inhibitors that are selective for mutant KRAS. Some of these inhibitors have been shown to be effective in patients with KRAS G12C-mutated cancers, and Sotorasib and Adagrasib have each received breakthrough designation from the FDA for the treatment of advanced or metastatic non-small cell lung cancer (NSCLC) harboring KRAS G12C mutations.
In 2022, the results of two important studies related to KRAS G12C, CodeBreaK100 and KRYSTAL-1, will be announced. The former study confirmed that the ORR of Sotorasib in pancreatic cancer was 21.1%, and the latter study confirmed that the ORR of Adagrasib in gastrointestinal cancer was 41%. Therefore, KRAS G12C has also obtained a new level3 rating for the corresponding cancer type in OncoKB.
What’s more interesting is that as KRAS becomes a hot spot, at the end of 2022, two large cohort retrospective analyzes of tens of thousands of people will be published in November and December, describing the population characteristics of RAS mutations from the perspective of Genomic Landscapes.
**If there is a sample database of hundreds of thousands of people, what can be done? **For today’s tweet, let’s take a look at the article published in npj Precision Oncology in December.

Main data and analysis content
The data of the article comes from Foundation Medicine (hereinafter referred to as FMI), and most of the authors come from FMI and Genentech.
In this study, the authors performed a comprehensive pan-cancer genome analysis to determine the frequency of KRAS mutations in 24 cancer types, including the distribution of G12C and mutations beyond KRAS.
Genomic signatures associated with different KRAS mutations were assessed by correlation analysis with TMB, PD-L1 expression, mutual exclusion concomitant and mutation signature.
In addition, the prognostic impact of different KRAS mutation subtypes in non-small cell lung cancer (NSCLC), colorectal cancer (CRC), and pancreatic cancer (PDAC) cohorts was assessed using clinical survival information collected in the FMI Clinical Genome Database (CGDB) .
As specific KRAS inhibitors and combination therapy strategies are developed, it is necessary to understand the correlation analysis of co-mutation and other biomarkers that may regulate targeting or immunotherapy . This is also the clinical value of this type of descriptive research.
Core conclusion
First look at the prevalence of KRAS in different cancer types
From December 2013 to December 2021, FMI tested a total of 426,706 tissue or blood samples from adult cancer patients (I don’t know how domestic tumor NGS colleagues feel about this number).
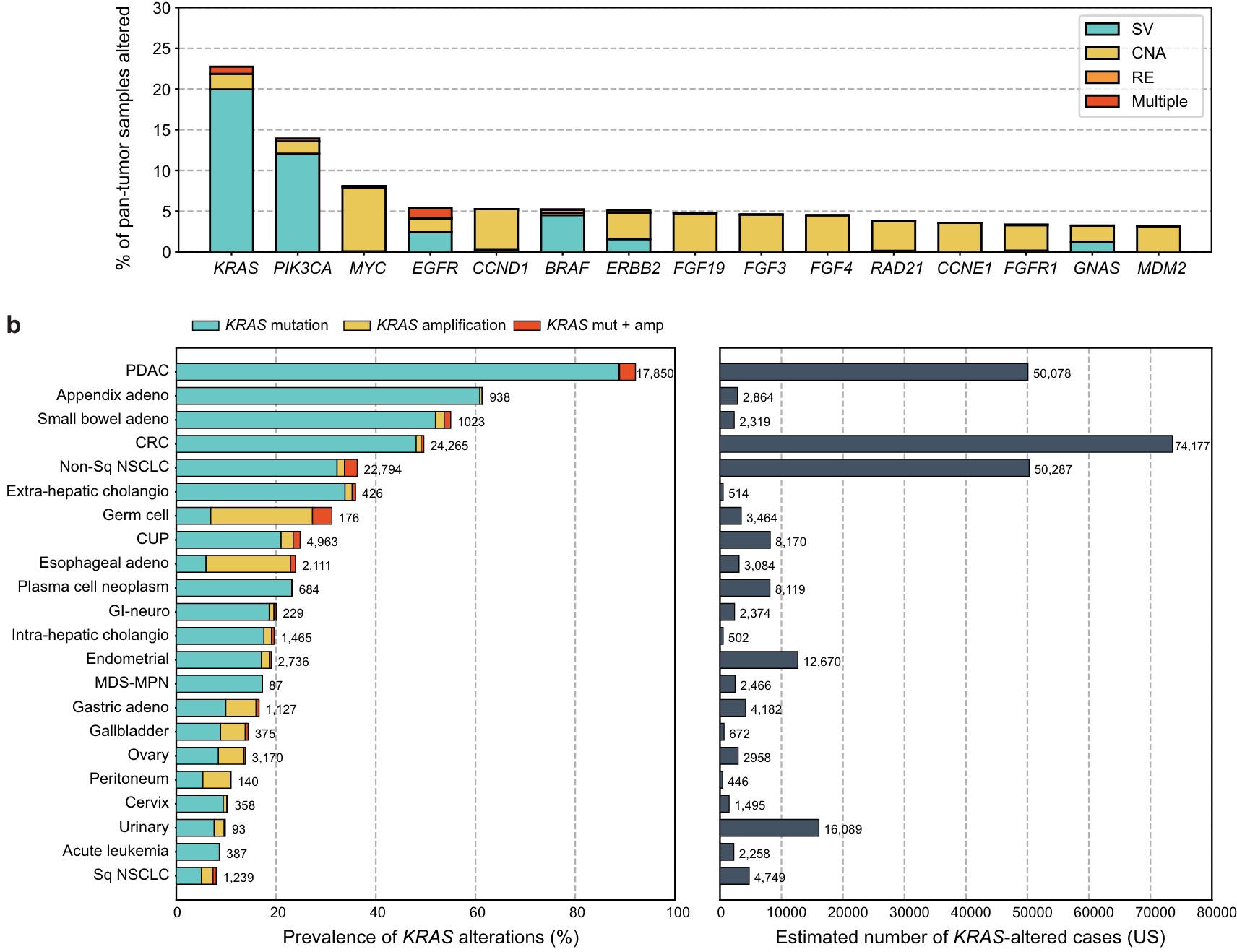
KRAS was the most frequently altered oncogene, with mutations found in 97,062 (23%) pan-tumor tissue samples, of which the vast majority (88%) were short variant mutations, 8.4% were amplifications, and 3.8% were mutations and amplifications. Increase accompanying hair.
Incidence was estimated from this prevalence data. In the United States, KRAS mutations are estimated to be highest in CRC with an estimated 75,000 cases, followed by PDAC and nonsquamous NSCLC. KRAS amplification was rare in most tumor types but was common in germ cell tumors (24%) and esophageal adenocarcinoma (18%).
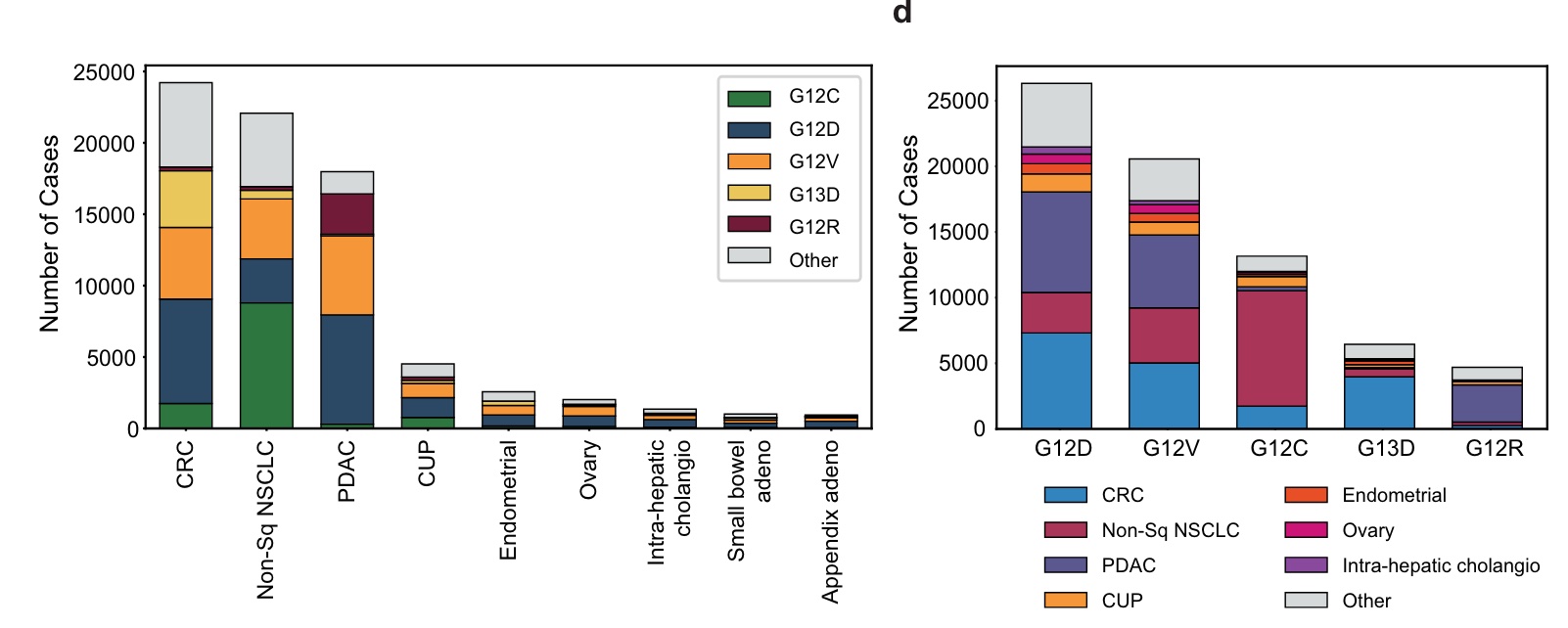
KRAS G12D (29%), G12V (23%), G12C (15%), G13D (7%), and G12R (5%) are the five most common KRAS mutations, accounting for approximately 80% of all KRAS mutations.
The tumor types with the highest incidence of KRAS mutation (KRASm) were PDAC (92%), appendix adenocarcinoma (61%), small bowel adenocarcinoma (SBA, 53%), CRC (49%) and non-squamous NSCLC (35%); CRC, nonsquamous NSCLC, and PDAC together accounted for 71% of the KRASm pan-tumor population.
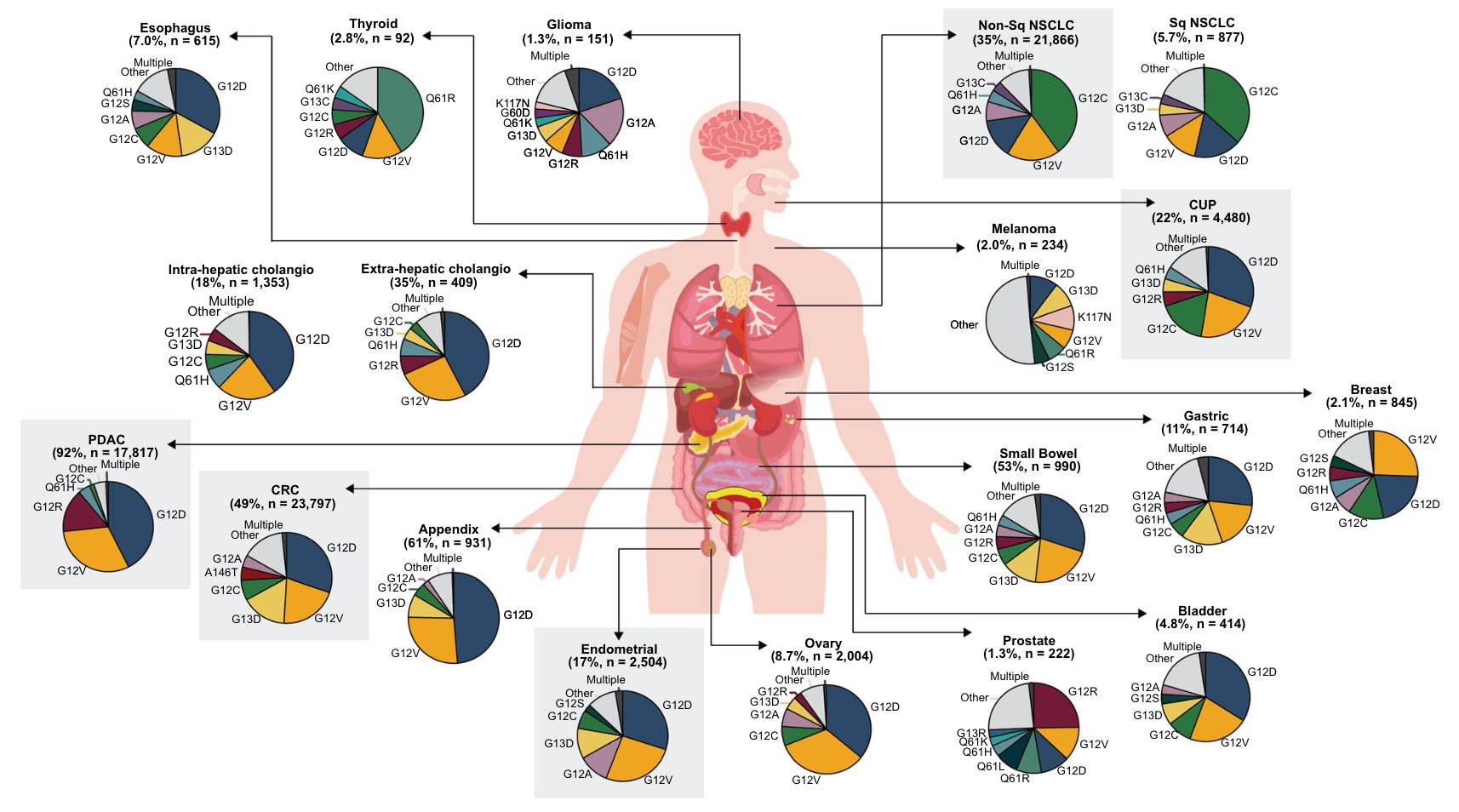
Although the distribution of KRASm varies among adult tumor types, similar KRASm patterns were observed in tumors from similar tissue types.
KRAS G12C was the most common variant in NSCLC (Non-Sq and Sq accounted for 40% and 36%, respectively).
Gastrointestinal tumors (including colorectal cancers as well as cancers of the esophagus, stomach, small intestine, and appendix) were similarly characterized, with KRAS G12D and G12V being the two most common variants.
KRAS G12D was also most common in many other tumor types, including PDAC (43%) and endometrium (30%), KRAS G12V was the second most common in most tumor types studied, and most common in breast cancer (26 %).
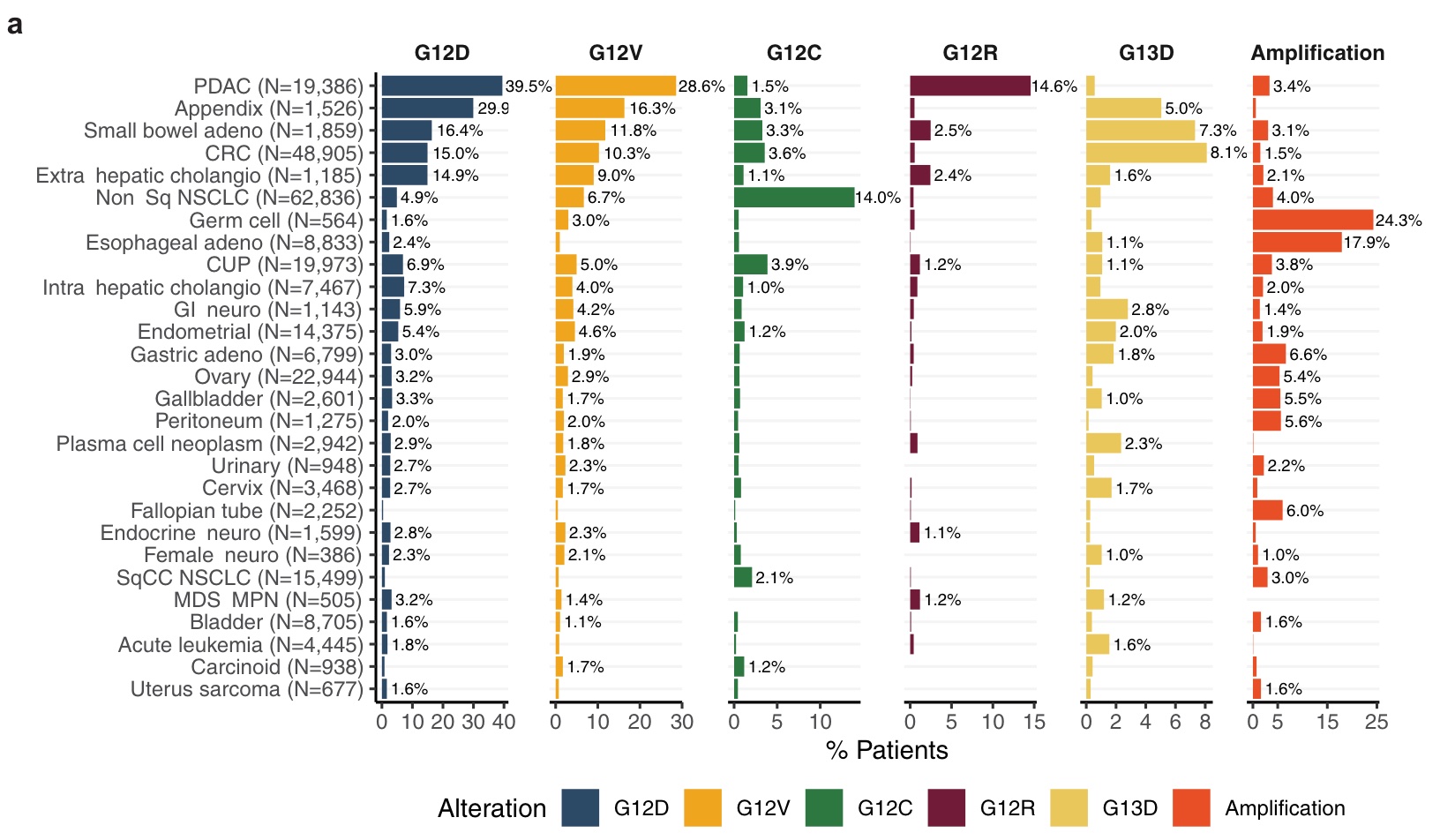
After knowing the incidence rate. What will be the characteristics of KRAS in co-mutation, mutation signature and immunotherapy biomarkers? Let’s look at the results of the analysis in this study one by one.
Across the four major KRAS-mutant tumor types, mutually exclusive volcanoes associated with KRAS are shown below. It is worth noting that in non-squamous NSCLC, colorectal cancer and endometrial cancer, TP53 gene is the most commonly mutated gene mutually exclusive with KRAS, while in PDAC, TP53 gene mutation is often associated with KRAS.
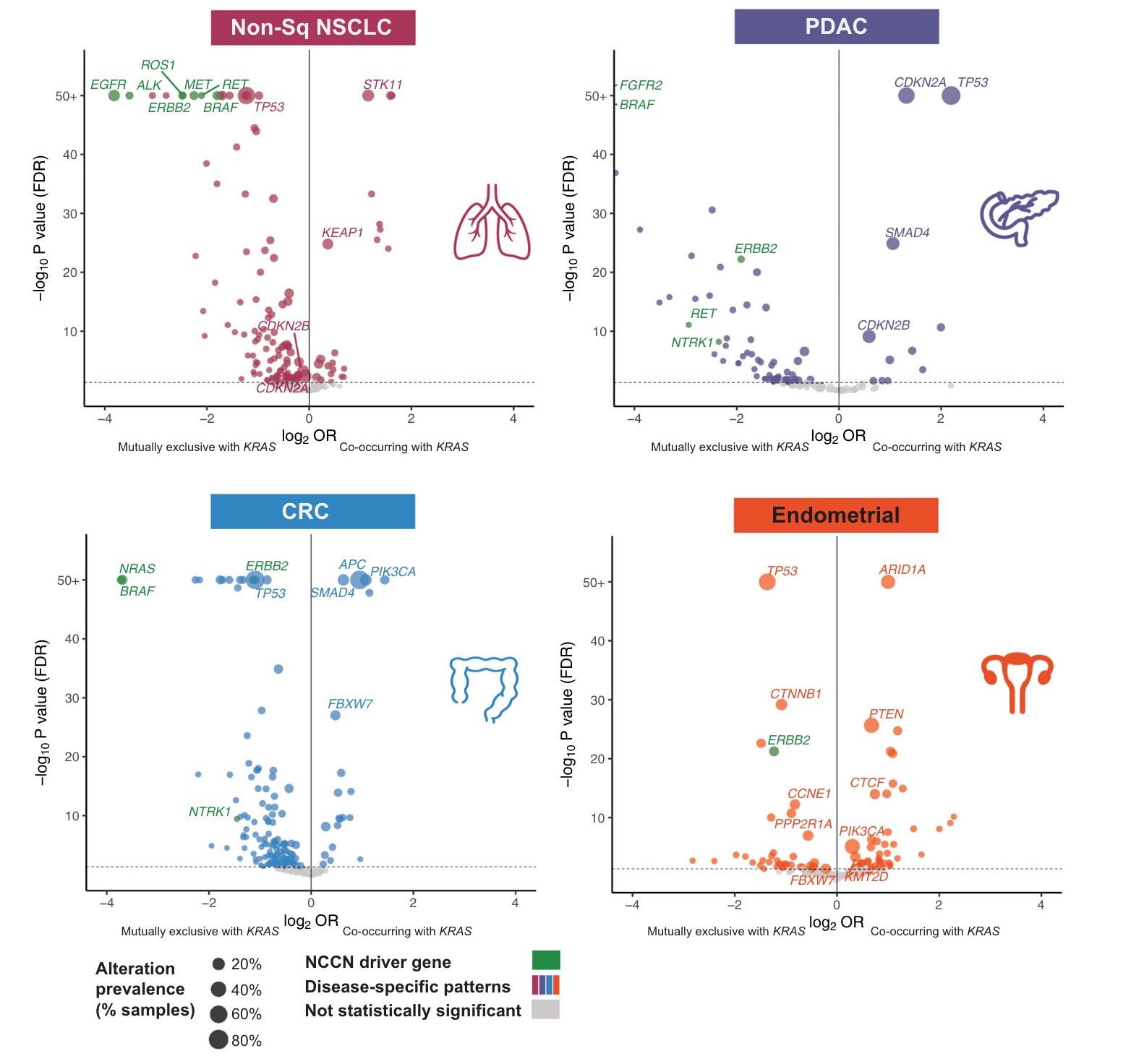
As a key driver gene, KRAS mutations are indeed highly mutually exclusive with mutations in other driver genes (EGFR, ALK, MET, ERBB2, BRAF, RET, and ROS1) in the RTK/MAPK pathway.

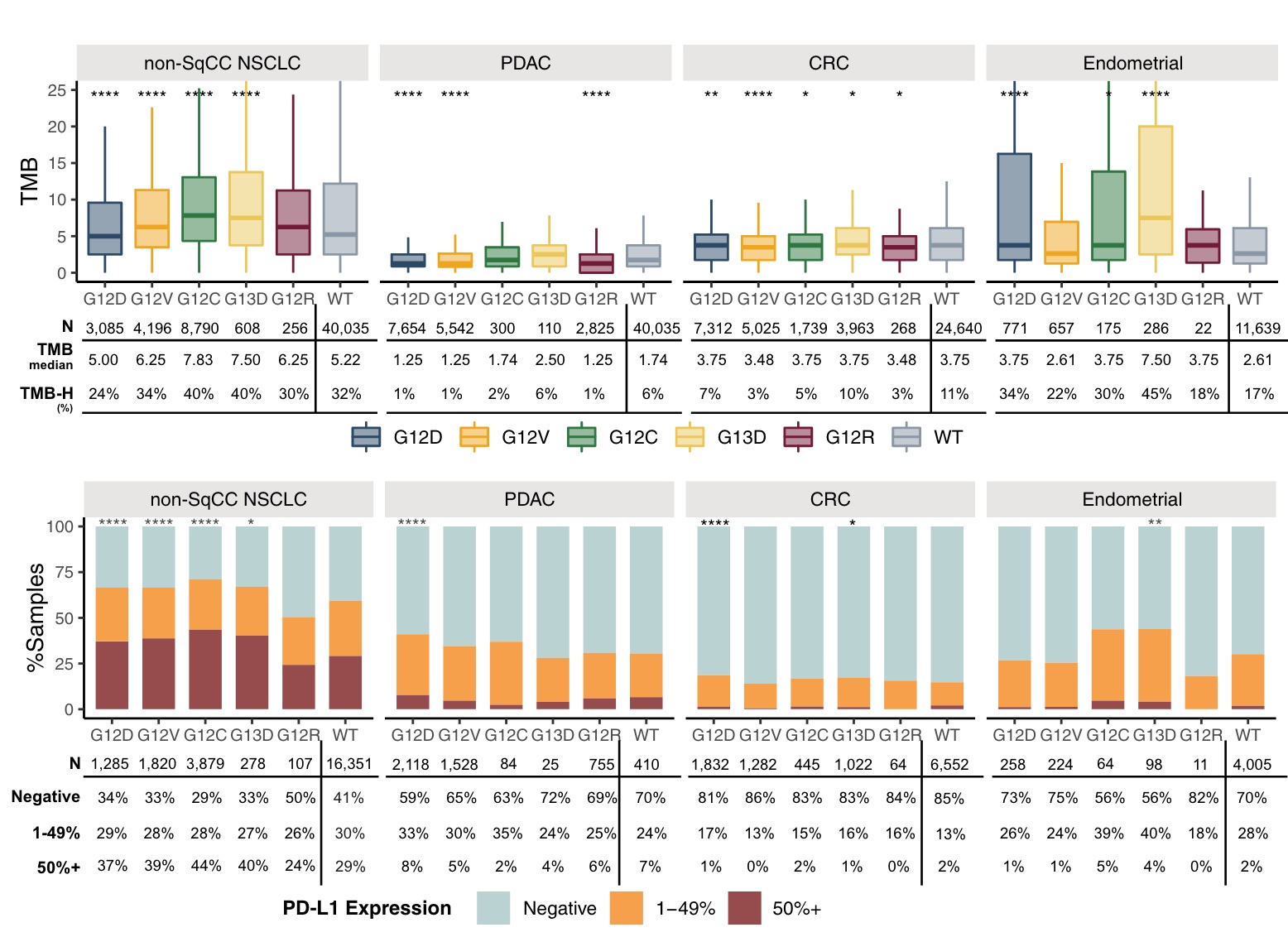
In nonsquamous NSCLC, KRASG12C-mutant tumors were enriched for high TMB and high PD-L1 expression, whereas KRASG12D-mutant tumors had a lower incidence of high TMB relative to other KRASm isoforms.
Relative to non-Sq NSCLC, KRASm had a lower proportion of positive samples with high PD-L1 expression in CRC, PDAC, and endometrium, whereas endometrium was similar to NSCLC, with 21–34% of TMB ≥10.
In endometrial carcinoma, KRAS G13D, G12D, G12C and G12A were enriched for high TMB compared with G12V and WT.
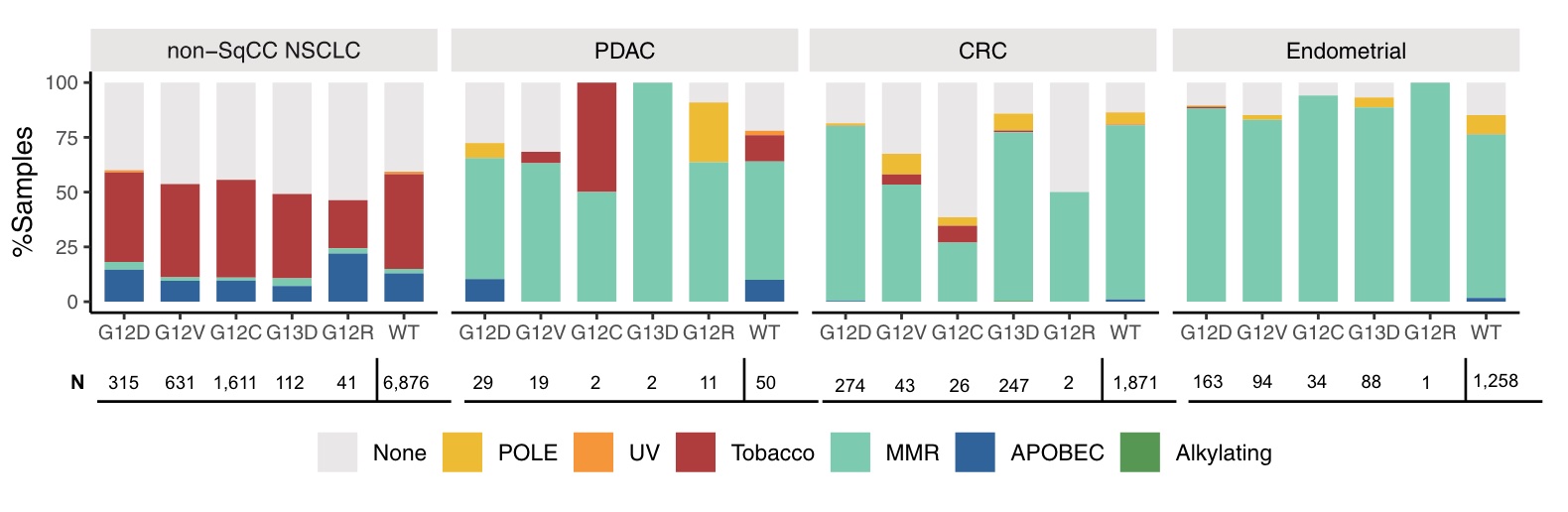
In addition, the mutation signatures corresponding to different mutation subtypes of four major cancer types were analyzed. Tobacco-associated features were detected in 38-45% of non-squamous NSCLC of different KRASm and KRAS WT subgroups, but less frequently (22%) in G12R non-Sq NSCLC and rarely in other tumor subtypes. Mismatch repair (MMR) features are rare in non-Sq NSCLC but common in KRASm and KRAS WT PDAC, CRC and endometrial cancer, but less frequently in KRAS G12C CRC relative to other subgroups Low. POLE signatures were detected in small subsets of PDAC, CRC, and endometrium.
Finally, let’s look at the association between clinical survival and KRAS mutations.
In lung cancer, patients with KRAS G12C mutations had similar overall survival (OS) to patients with other common KRAS mutations, including G12V (11 vs. 10 mos, HR 1.0, 95% CI 0.86-1.20, p = 0.88 ) and G12D (11 vs. 12 mos, HR 0.91, 95% CI 0.75-1.11, p = 0.36), compared with the rarer non-G12/13 KRASm (11 vs. 9 mos, HR 1.1, 95% CI 0.82-1.35 , p = 0.67) OS was similar. NSCLC patients without KRAS mutations but exhibiting other oncogenic drivers had significantly better OS than NSCLC patients with KRAS G12C mutations. The results of univariate and multivariate analyzes were similar.
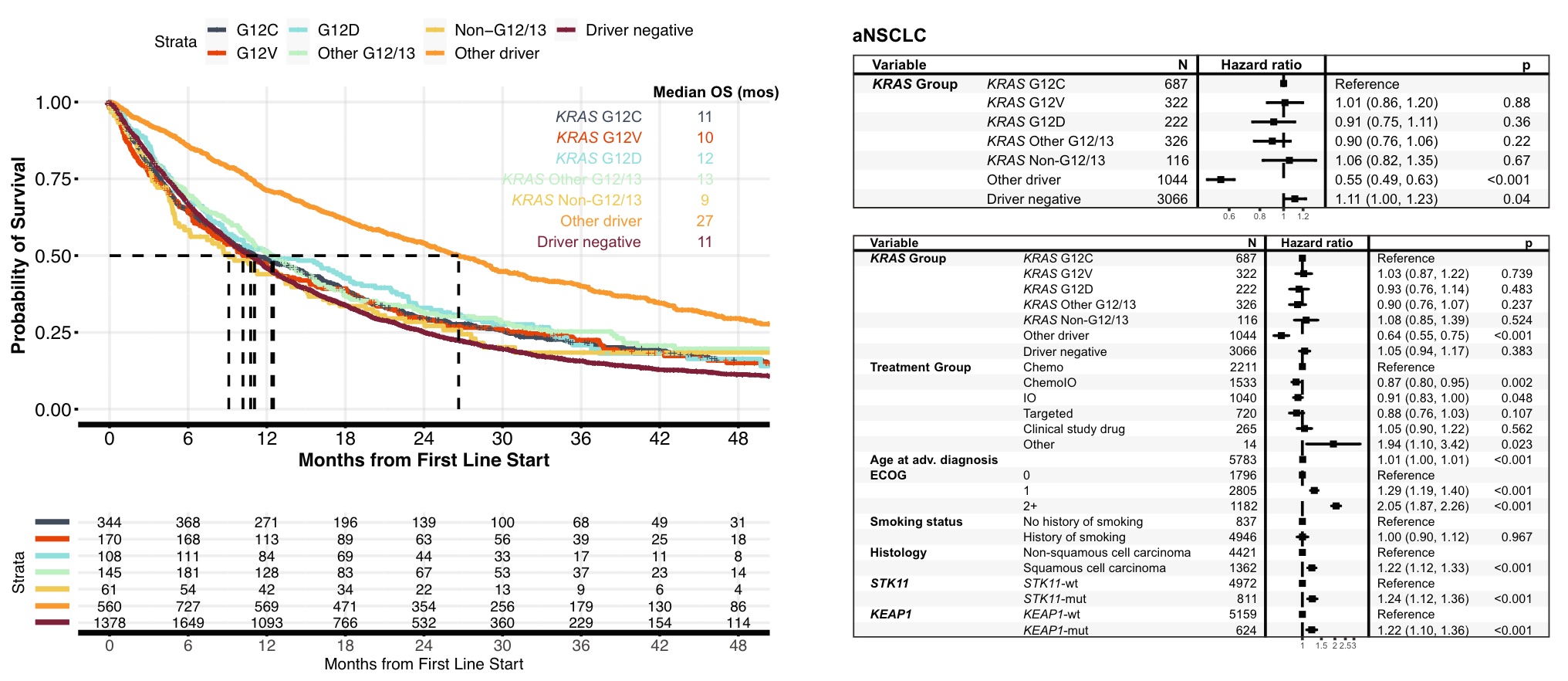
In CRC, patients with KRAS G12C mutations had similar OS to patients with other common KRAS mutations, including G12D and G12V. Compared with BRAF V600E patients and non-G12/13 mutation patients, patients with KRAS G12C mutation had a slightly higher OS, but not statistically significant. KRAS/NRAS/BRAF V600E-negative (RAS/RAF-negative) patients had a more favorable OS than the KRAS G12C subgroup (24vs. It is also consistent in the model
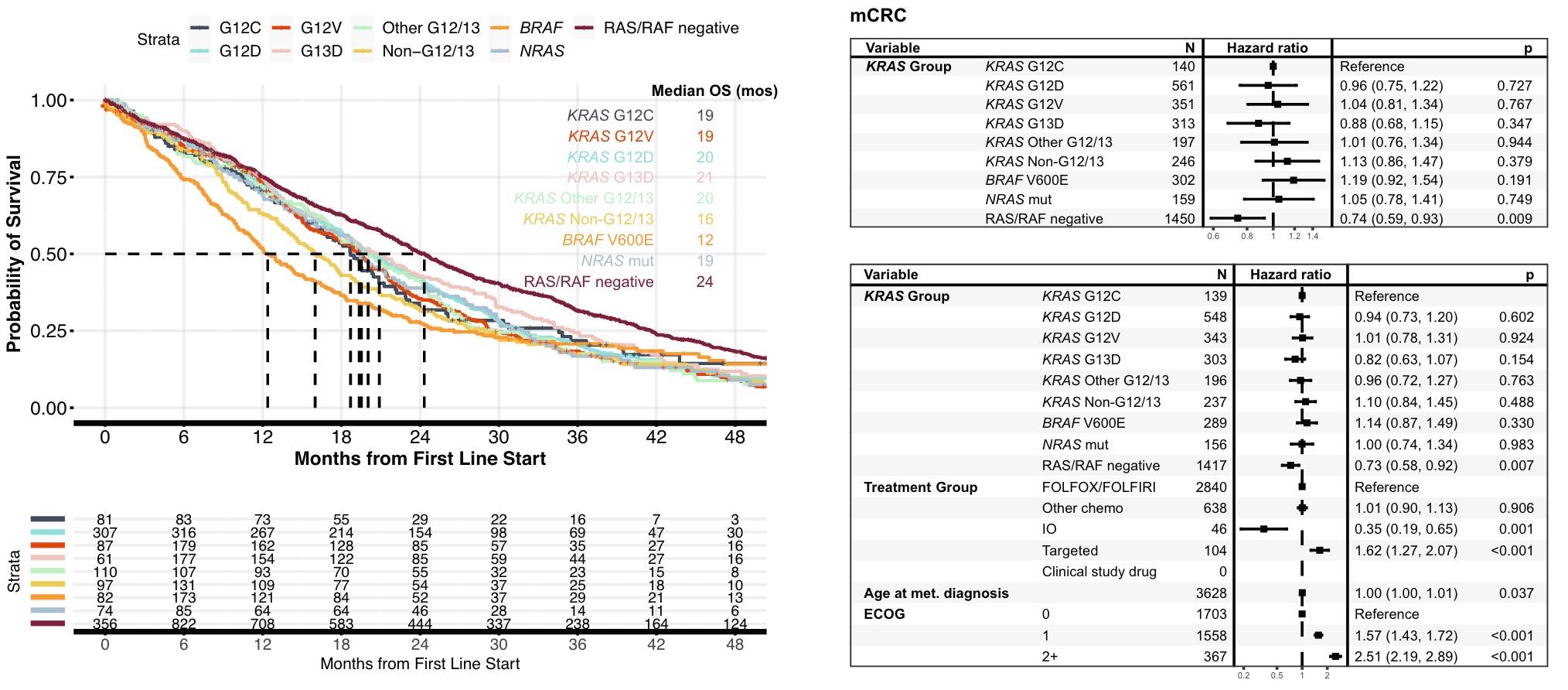
KRASm was found in 93% of metastatic PDAC patients, 94% of which occurred within codons G12/G13. OS in patients with tumors with KRAS G12C mutation was significantly lower than that in patients with KRAS-negative tumors (5.9 vs 8.8 mos, HR 0.59, 95%CI 0.37~0.94, p=0.03), and similar to that in patients with non-G12/13 KRASm tumors (5.9 vs 6.9 mos, HR 0.80, 95% CI 0.50~1.29, p=0.36). However, in a multivariate model, OS was not significantly worse in patients with KRAS G12C-mutant tumors compared with patients with KRAS-negative tumors, although this may be limited by the small number of KRAS-negative cases
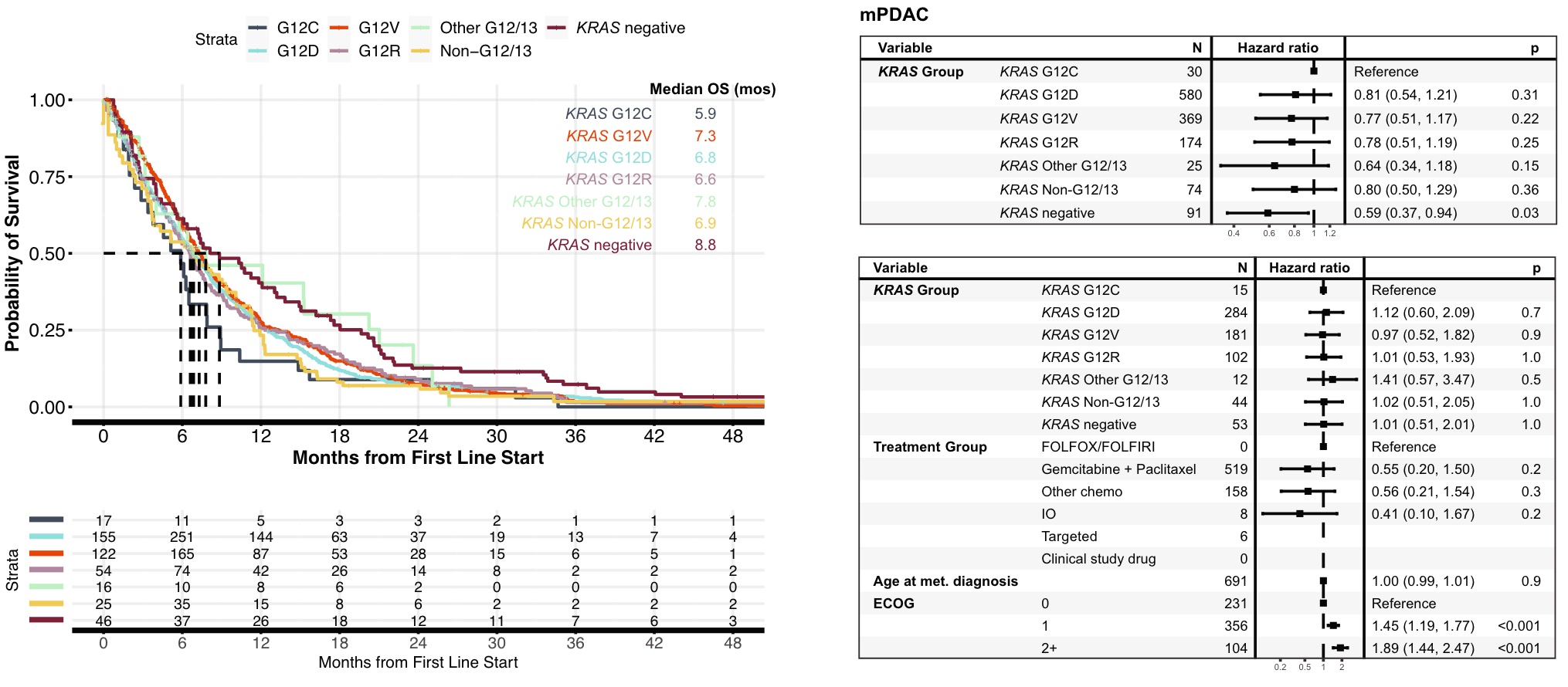
Despite these limitations, the findings of this study have important implications for the development of KRAS inhibitors targeting G12C, G12D, G12V, and higher levels. Genomic profiles that detect co-alteration and mutational signatures, as well as trials that understand the clinical importance of these biomarkers as predictors of response to targeted therapy and immunotherapy, will be imperative to improve treatment selection and outcomes.
say a few more words
The method part of this article is very time for practitioners in related industries to understand what different FMI products can be used for, and roughly how to do it. Many details of the analysis are worthy of careful consideration and understanding~
As the absolute leading company in international tumor NGS detection, FMI has accumulated a large number of samples in its own data sample library. As long as there is a breakthrough in the clinical results of a certain gene-corresponding drug, or important functional research results in tumor-related research, they can quickly conduct large-scale analysis in hundreds of thousands of sample libraries.
Considering the length of this article, similar studies will not be listed one by one. If you are interested, you can search and search. Later, an integrated topic introduction will be introduced in the membership plan of the Xiongyan Xiongyu email newsletter . Welcome to scan the QR code below to subscribe for free, and receive content updates as soon as possible.
The author of this article : bear who thinks about problems
Copyright statement : Unless otherwise stated, all articles on this blog are licensed under the Creative Commons Attribution-Non-Commercial Use-No Derivatives 4.0 International License Agreement (CC BY-NC-ND 4.0) .
If you are interested in this article, please subscribe to my “Xiong Yan Xiong Yu” member newsletter via email or WeChat. I will share with you the latest industry research progress in the field of tumor biomedicine and what I think and learn. If you want , click this link to subscribe for free. 
This article is transferred from: https://kaopubear.top/blog/2022-12-16-fmi-kars-pancancer-landscape/
This site is only for collection, and the copyright belongs to the original author.