Original link: https://kaopubear.top/blog/2022-08-02-ctdna-esmo-caca-recommendation/
write in front
In June 2022, the Professional Committee of Tumor Markers of the Chinese Anti-Cancer Association (CACA) published the Expert Consensus on Clinical Practice of ctDNA High-throughput Sequencing (2022 Edition) in the Chinese Journal of Cancer Prevention and Control.
In July 2022, the European Society for Medical Oncology (ESMO) Precision Medicine Working Group published recommendations for ctDNA testing in cancer patients at the Annals of Oncology .

At the same time, on July 20, 2022, Nature released a related study on high-depth whole-genome ctDNA sequencing in prostate cancer, revealing the potential application direction of ctDNA in the future from another dimension.
Deep whole-genome sequencing of ctDNA can reveal unique characteristics of each patient, helping clinicians to more effectively detect treatment resistance and select personalized treatments that can improve patient outcomes.

Subsequently, on August 1, 2022, Nature Reviews Clinical Oncology published another ctDNA review: Circulating tumour DNA — looking beyond the blood, which introduced the current status of ctDNA applications in non-blood sample types.

Considering the space problem, we will complete the study of these series of articles in three times. The first article is the sorting and study of the consensus recommendations of ESMO and CACA experts; the second and third articles are the interpretation study of the two literatures mentioned above. .
It is hoped that through these three articles, we can have a relatively comprehensive understanding and grasp of the current technical problems, application directions and new application prospects of ctDNA.
Note : In this article, we will weaken the specific experimental and technical details of the consensus, and focus on the content of the two consensuses involving ctDNA biological characteristics, technical analysis issues, and clinical application directions. For more complete information, consult the original documentation. The overall structure of this paper is based on the clinical application recommendations of ESMO, and cooperates with the corresponding CACA expert consensus.
ctDNA biological features
In healthy people, plasma DNA is mainly derived from cells of the hematopoietic system, with concentrations ranging from negligible amounts to as high as 100 ng DNA/ml, and plasma DNA fragments produced by normal cells are around 166 bp in length. The DNA fragment (ctDNA) actively secreted by tumor cells or released into the circulation during tumor cell apoptosis or necrosis is about 132-145 bp in length, and has a short half-life (usually <2 h)
ctDNA carries tumor cell-related genetic features, including mutation, methylation, amplification and rearrangement. ctDNA length, genomic location, and epigenetic markers, among others, can provide information to differentiate normal and cancerous samples and to determine the origin of tumors. ctDNA represents the mixed DNA released by different tumor subclones and can reflect specific tumor heterogeneity and better characterize tumor genomes.
Plasma DNA clearance mainly occurs in the liver, and the factors affecting the dynamic changes of ctDNA levels mainly include the following:
-
Tumor histological type, location, stage, tumor burden
-
DNA interference from other sources
-
Gene mutation information carried by cfDNA produced by clonal hematopoietic cells
-
different sampling times
-
medical treatement
ctDNA is a DNA fragment actively secreted by tumor cells or released into the circulatory system by tumor cells during apoptosis or necrosis; its abundance is affected by many factors and fluctuates greatly. Biological information may evolve and be interfered with by normal cell germline mutations or clonal hematopoietic cell line mutations , and special attention should be paid during clinical testing and report interpretation. (CACA Consensus)
ctDNA detection technical problems
In cancer patients, ctDNA varies according to different characteristics of the tumor, including tumor location, disease extent, proliferation and apoptosis, necrosis, inflammation, tumor microenvironment, and more. The key to the success of ctDNA assays is the selection of the appropriate assay type to address specific scientific and clinical questions.
Currently, there is no single ctDNA analysis method that is suitable for all purposes (early detection, MRD, gene mutation identification and assessment of tumor genetic heterogeneity, identification of molecular mechanisms of resistance, etc.).
Ultrasensitive analysis of limited amounts of ctDNA is required in the context of early detection or MRD analysis, while different types of analysis are required to identify mutations associated with treatment resistance and tumor genetic heterogeneity in the metastatic setting. Different assays have different degrees of LoD, LoQ, which ultimately determine the amount of plasma DNA and ctDNA required to provide analytical results.
Regarding the technical issues of ctDNA, we need to focus on the following aspects:
Impact before analysis
The release of ctDNA is influenced by many factors, such as patient-specific physiological conditions, inflammation, and acute and chronic diseases. The level of ctDNA is affected by various treatment modalities including chemotherapy, targeting and immunization, and can be divided into two stages: acute changes and long-term changes.
Among these, acute changes ranging from days to weeks are directly affected by treatment on tumor and normal cells, and long-term dynamic changes from weeks to months are associated with tumor shrinkage by treatment.
In addition, the amount of plasma DNA available for detection is proportional to the volume of plasma extracted, and care must be taken to avoid lysis of leukocytes during sample collection and processing.
Impact in Analysis
During analysis, false positives and false negatives are the two main problems.
Even in relatively advanced cancer patients, many plasma ctDNA levels may be low. Undetectable mutations in ctDNA testing may be due to the absence of the mutation in the patient’s tumor (true negative) or the low level of ctDNA to detect the mutation (false negative).
Various technical factors need to be considered in terms of the causes of false negative test results, such as the amount of plasma DNA analyzed and possible differences in detection sensitivity between different types of mutations. Plasma ctDNA concentration correlates with tumor stage and volume, and several clinical and pathological factors also correlate with ctDNA levels. Interpretation of ctDNA test results must take into account the possibility that plasma samples do not contain sufficient ctDNA to detect different types of mutations.
When using CNAs, DNA methylation, or plasma DNA fragment analysis, the likelihood of giving a true negative result needs to be increased by ensuring that sufficient tumor-derived DNA is present. The figure below shows the assays that can be performed at different ctDNA levels.
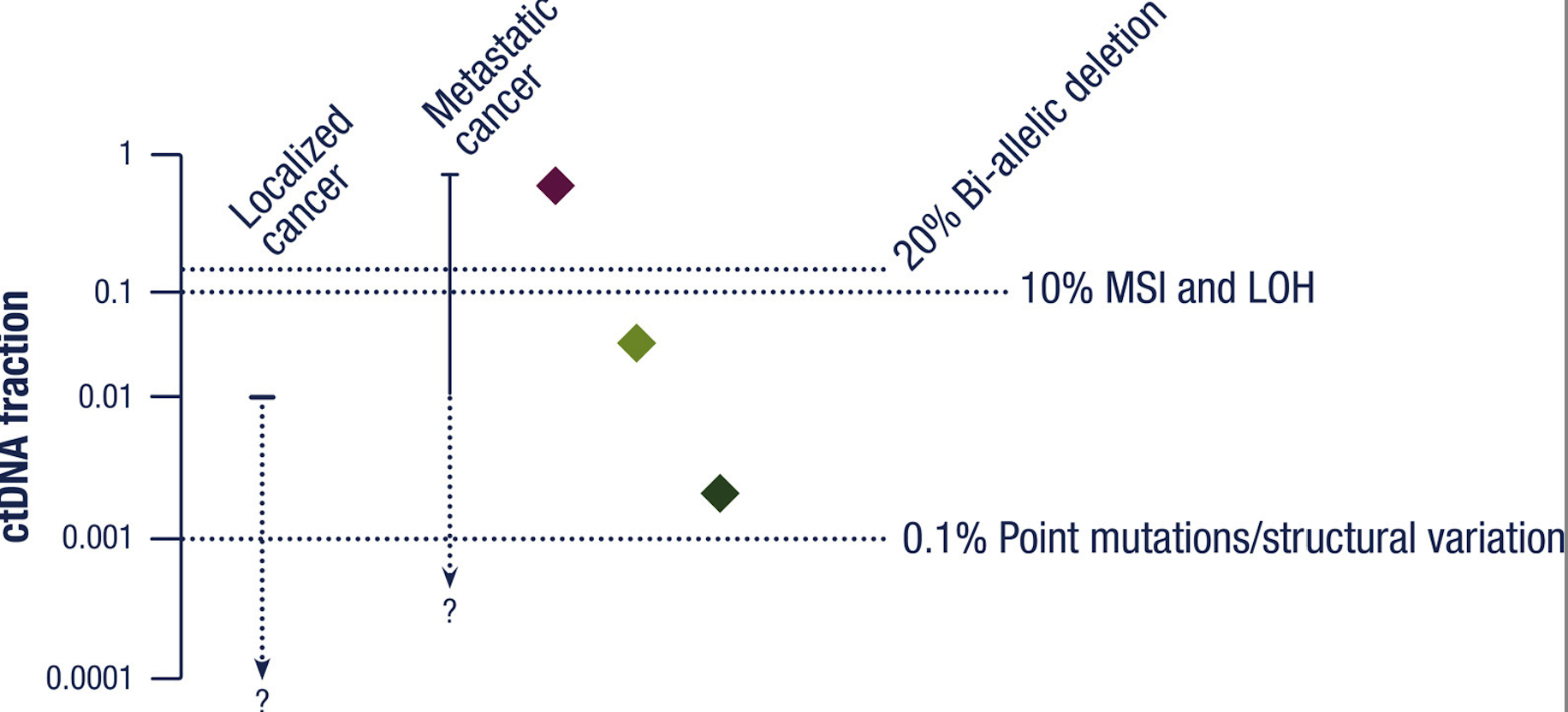
For false positive results, the biggest problem comes from clonal hematopoiesis and germline mutations.
Somatic alterations accumulate in normal tissues throughout the cell life cycle due to factors such as environmental, chemical factors or replication errors during cell division. If these somatic alterations confer a selective growth advantage, it may cause clonal expansion and increase the probability of successive driver mutations. Plasma DNA in blood is primarily derived from apoptotic hematopoietic cells, so clonal expansion of these cells may lead to false-positive results for ctDNA.
Genes mutated in clonal hematopoiesis (CHIP) partially overlap with solid tumor driver genes. For example, a large proportion of patients with advanced prostate, lung, and breast cancers have been detected with false positives for clonal hematopoiesis-related genes (eg, TP53, ATM) in plasma DNA. These false positives can be largely ruled out by leukocyte (WBC) DNA sequencing or by paired samples of tumor tissue samples.
It is therefore important to note that plasma-only DNA analysis of many tumor suppressor genes (eg, TP53) or genes involved in DNA repair (ATM and CHEK2) is challenging, especially for mutated genes commonly found in clonal hematopoiesis, such as DNMT3A and TET2. Be careful.
In response to these problems, ESMO’s expert advice provides the following technical suggestions that need to be considered.
-
The timing of blood draw for ctDNA analysis should be carefully chosen based on the clinical question, with different factors affecting ctDNA release (eg, therapy, concurrent inflammatory processes, and surgery). To detect postoperative MRD, it is best done within at least 1 week after surgery; for major surgery with a longer healing time, it may take 2 weeks or more. For advanced cancer genotyping, sampling during treatment response or non-progression should be avoided to minimize false negatives.
-
Depending on the processing time and the assay method used, care should be taken when selecting blood sample collection tubes.
-
If blood samples are collected prior to DNA extraction, they should be stored at -80°C for long periods of time to minimize continuous freeze-thaw processes.
-
False negatives (relevant mutations actually present in the tumor not being detected) are a significant problem with ctDNA testing and can be due to low levels of plasma DNA analyzed, insufficient detection sensitivity, or “non-shedding” tumors .
-
Clonal hematopoiesis is a common cause of false positives in ctDNA assays when testing for genes prone to contain clonal hematopoietic mutations. For the detection of clinically accessible tumor suppressor genes such as DNA repair genes, it is recommended to analyze both plasma DNA and leukocyte DNA. For this assay, routine collection of leukocyte-enriched Buffy coats from patients undergoing plasma ctDNA testing is recommended so that material is available to rule out relevant effects if necessary.
-
Pathogenic germline mutations in tumor susceptibility genes (eg, BRCA1, BRCA2, PALB2) may be detected in ctDNA, and detection of such mutations requires the use of validated assays to determine somatic or germline.
-
Clinical genotyping assays should be adapted to assess tumor purity in the future to help physicians make true negative predictions.
For the relevant standards of ctDNA NGS detection, the corresponding CACA consensus is as follows.
The quality management of ctDNA NGS testing laboratories needs to run through the whole process, and the ctDNA collection, sample processing and automation processes should be carried out in accordance with standardized and clinical validation procedures to minimize the possibility of false negatives caused by operational differences. It is recommended to use anticoagulant tubes containing cell stabilizer for sample collection, and complete plasma separation as soon as possible. It is recommended that the extracted cfDNA be tested within 24 hours. Otherwise, store at -30°C to -15°C and avoid repeated freezing and thawing. (CACA Consensus)
The technical route of ctDNA NGS detection should be selected according to the needs of the project, and different sequencing strategies can be selected according to the number of genes detected and the coverage. When performing ultra-sensitive mutation detection based on ctDNA, it is recommended to use molecular tagging technology and optimize the corresponding bioinformatics analysis settings to reduce false positive results due to random errors of the sequencing platform; it is recommended to establish sequencing noise and clonal hematopoietic background libraries. This method reduces the effects of clonal hematopoiesis and background noise. (CACA Consensus)
Test report information
ESMO has made the following recommendations on what should be included in the test report and the wording specification.
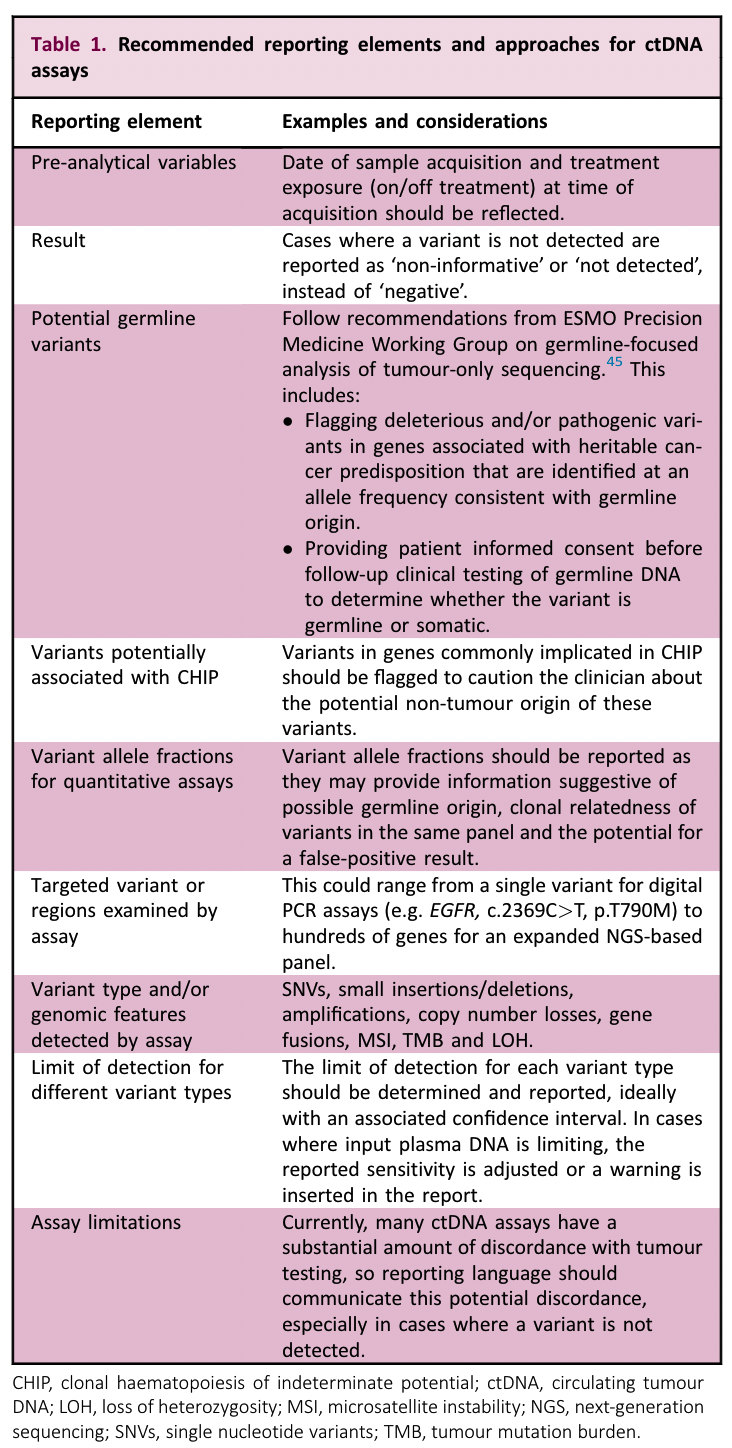
The CACA consensus mentions:
The ctDNA NGS clinical test report should include the basic information of the subject, sample information, laboratory information, test items, test results and interpretation of variants, the results of internal laboratory validation of the test method, test limitations and uncertainties, and recommendations for further testing and so on. The laboratory should establish a reporting SOP, and it is recommended to classify or grade the detected tumor gene mutations according to domestic and foreign literature, consensus guidelines, clinical trial evidence and practice. (CACA Consensus)
If you want to know more comprehensive content, you can refer to another Chinese consensus on the interpretation of clinical report on next-generation tumor sequencing .
Clinical application of ctDNA detection
For the clinical application of ctDNA, ESMO gives the following paragraph description.
ctDNA has a variety of potential clinical applications, including for screening, characterization of early stage disease, detection of molecular residual disease (MRD) after local therapy, prediction of recurrence, classification of advanced tumors, early assessment of treatment effect, response monitoring and identification of resistance mechanisms. (As shown below)
ctDNA has multiple potential clinical applications, including its use for screening, characterisation of early disease, detection of molecular residual disease (MRD) after definitive local treatment, prediction of relapses, genotyping advanced cancer, early assessment of treatment efficacy, monitoring of response and identification of mechanisms of resistance to therapy
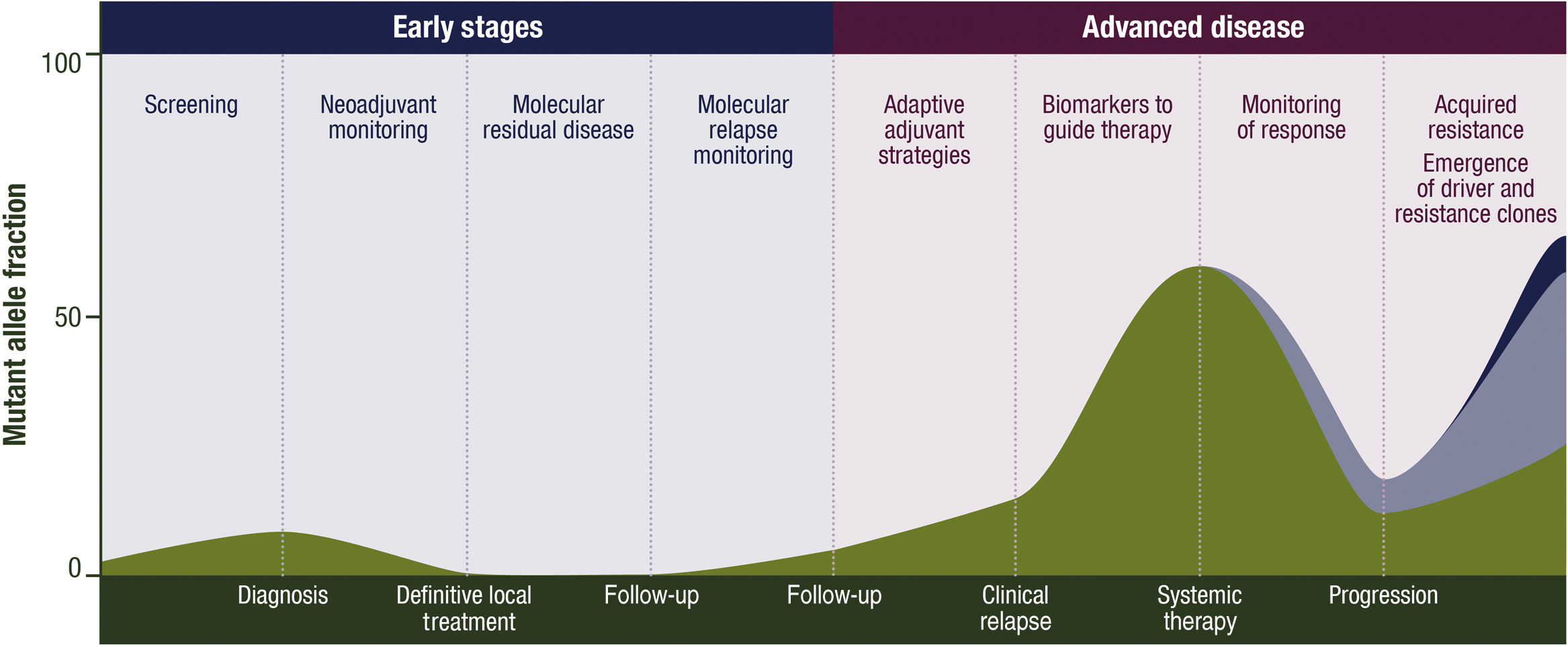
For patients requiring urgent treatment, delays in tissue biopsies can limit treatment options, while liquid biopsies can provide faster results.
Spatial and temporal heterogeneity is an established feature of malignancies, and ctDNA has the advantage of being more readily available serially and obtaining a “genomic repertoire” from several metastatic sites in a patient. When tissue testing can only be performed in the primary tumor, liquid biopsies can provide more accurate genotyping for patients with metastases.
The release of ctDNA is proportional to tumor growth, which is related to cell death and turnover. The fastest growing tumor clones shed the most ctDNA, which is theoretically the most clinically significant. Multiple liquid biopsies may allow detection of acquired resistance mutations for optimal selection of subsequent therapy based on acquired resistance genotype.
Compared to tissue testing, ctDNA testing also suffers from higher false-negative and false-positive rates, and a lower proportion of tumors in the sample, limiting accurate assessment of mutant allele fraction (VAF) and limiting copy number variation analysis.
Advanced tumor genotyping
The following table lists recommendations for tumor-specific advanced tumor genotyping testing for liquid biopsy.
For specific content, please refer to the original text and the corresponding treatment guide.
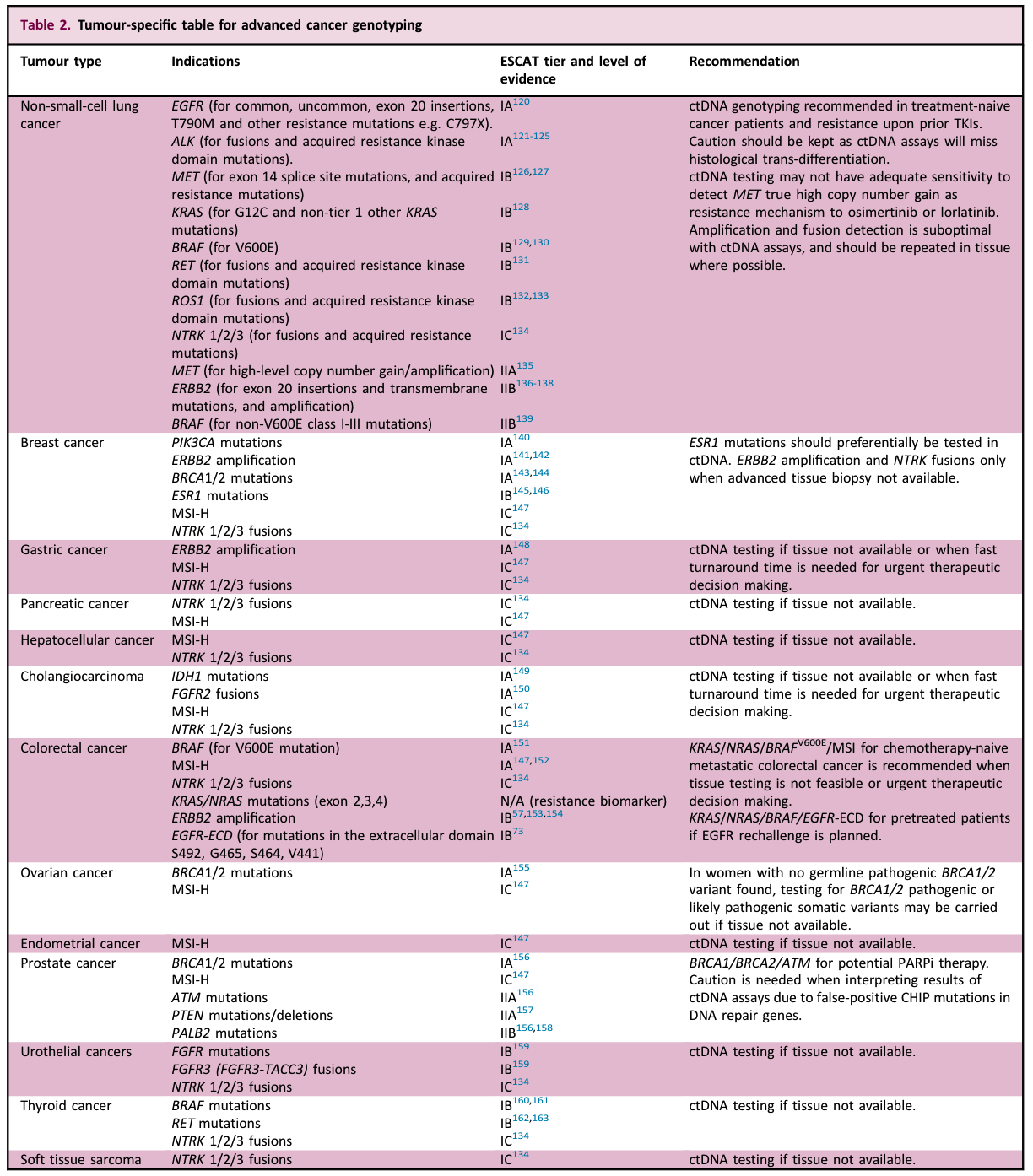
Even though there is abundant evidence that liquid biopsy can be used for advanced tumor genotyping, it still has obvious limitations.
For different mutation types, the performance of ctDNA detection is still very different. For example, somatic copy number variation in ctDNA samples can only be accurately identified when the proportion of ctDNA is high, and can only replace tissue assessment when tissue sampling is not possible.
In addition, bTMB is highly correlated with the amount of ctDNA, so a minimum amount of ctDNA is required for a valid evaluation (this issue also applies to tissue testing when tumor cell content is low)
Recommendations for tumor testing sites
Sensitivity of ctDNA assays is reduced in CNS-only metastatic disease and primary brain tumors, for which liquid biopsy is generally not suitable for genotyping, but may be attempted if it is the only sample source for genotyping .
Studies have shown that genotyping can be obtained from cerebrospinal fluid analysis, but in the presence of oligometastasis and nodular-only disease, ctDNA has a high false-negative rate, and special consideration should be given to tissue testing or interval testing in larger tumors ctDNA detection.
Recommendations for the application of advanced tumor genotyping
-
Liquid biopsy has very high sensitivity and clinical specificity in advanced tumors and can be used in routine practice when its results would affect standard treatment options. The limitations of ctDNA detection must also be considered.
-
Liquid biopsy is recommended as an alternative to tissue genotyping, especially for aggressive tumors where rapid results are clinically required, such as advanced NSCLC. This also applies when tissue biopsy is not possible or appropriate.
-
Liquid biopsy for genotyping should be performed at tumor progression (whether untreated or later-line therapy). The sensitivity of the tumor is reduced when samples are collected when the tumor is responding to treatment.
-
For genotyping of advanced tumors, in clinical practice, the choice between RT-PCR, dPCR, and NGS testing should be based on availability, reimbursement, and the number of grade 1 actionable genetic variants in the tumor-specific setting.
-
Caution should be exercised when interpreting pathogenic variants in cancer susceptibility genes (eg, BRCA1, BRCA2, PALB2), and effective germline testing is required to confirm germline or somatic mutations.
-
Given the clinical sensitivity and negative predictive value, tissue testing should be recommended when liquid biopsy does not detect a treatable mutation. For experts, tissue testing may not be necessary if sufficient ctDNA can be confirmed in the sample.
-
ctDNA assays are less sensitive for fusions and copy number variations, and tissue assays should be used when tissues are present.
-
All oncologists should have access to MTB, study before using ctDNA testing to ensure correct interpretation of test results, and discuss difficult cases to ensure sound decision-making.
Advanced tumor monitoring
Dynamic analysis of ctDNA for early efficacy assessment
Because of the short half-life of ctDNA and the potential for non-invasive repeat sampling, blood ctDNA can provide real-time monitoring of disease during treatment.
Studies monitoring tumor patients during treatment suggest that ctDNA dynamics correlate with treatment response and may be detected earlier than clinical/radiological testing. Across many different tumor types and types of treatment (chemotherapy, targeted, and immune), patients who responded to treatment experienced a decline in ctDNA levels within a few weeks of starting treatment.
The early decline in ctDNA may reflect reduced ctDNA release due to cell cycle arrest, and the later decline in tumor volume. It is important to note that there may be a brief rise in ctDNA levels a few days after starting cytotoxic drug therapy.
There is growing evidence that tracking changes in serial ctDNA levels in patients with metastatic tumors treated with immune checkpoint inhibitors can assess prognosis and treatment efficacy. Among them, one clinical use of monitoring ctDNA levels is to distinguish true clinical radiological progression from pseudoprogression of immunotherapy (which occurs in 5%-10% of patients receiving immunotherapy).
Longitudinal surveillance for detection of drug resistance mutations
ctDNA analysis can also be used to assess the emergence of genomic mechanisms of drug resistance prior to clinical progression. Several studies in cohorts of patients with different tumor types receiving targeted therapy have demonstrated that longitudinal ctDNA profiling is able to detect early-emerging resistance mutations before clinical progression.
future research directions
In the future, further studies are needed to determine the optimal timing of ctDNA dynamic assessment and accurate thresholds for predicting response. Further research is needed on the frequency of surveillance for drug-resistant mutations that appear before clinical progression.
Randomized intervention studies are needed to assess whether changing treatment based on dynamic assessment of ctDNA can improve patient outcomes or improve quality of life by avoiding unnecessary side effects and minimizing economic costs.
Recommendations for Advanced Tumor Surveillance
There is insufficient evidence to warrant periodic monitoring of ctDNA during treatment. Although early monitoring of ctDNA dynamics is strongly associated with prognosis, and resistance mutations can be identified many months before clinical progression, there is insufficient evidence that actions based on these findings can improve prognosis. Clinical trials with randomized interventions are needed to assess the effects of ctDNA surveillance.
Early tumor MRD and molecular recurrence monitoring
There is substantial evidence that the detection of ctDNA after potentially curative treatment is associated with a higher risk of future recurrence, and we generally use two terms to describe it. MRD is a term in solid tumors that describes the molecular evidence of residual cancer cells that appear shortly after curative treatment of the primary tumor with surgery and/or chemoradiotherapy.
MRD can only be detected by molecular techniques such as PCR or NGS or CTC analysis, but not by blood-based protein tumor markers or routine imaging tests. Molecular relapse (MR) refers to molecular detection of recessive disease at a later time point during adjuvant therapy or surveillance.
Data from multiple case-control and cohort studies support the clinical validity of ctDNA analysis for MRD detection. In many studies of early-stage breast, colorectal, lung, and bladder cancers, ctDNA detection immediately after completion of treatment or during surveillance can predict high-risk recurrence.
ctDNA testing showed that the specificity for predicting relapse in the absence of further treatment was often greater than 90%, however in most studies the sensitivity for detecting MRD shortly after completion of treatment was often less than 50%.
In addition, ctDNA testing shows that the time from ctDNA detection to clinical relapse is usually 6 months, which means that existing ctDNA MRD testing mainly detects those patients who are destined to experience early relapse (relapse within the first year after initial treatment), which may be Late recurrence of disease cannot be detected.
In addition to clinical efficacy, the clinical utility of ctDNA MRD remains to be determined and prospective randomized trials are needed. The most obvious potential application of MRD testing is to guide individualized adjuvant therapy, where MRD-positive patients may benefit from additional therapy. In addition, in theory, patients without MRD can reduce the intensity of adjuvant therapy. However, the currently available ctDNA MRD assays are not sensitive and have a high false-negative rate. The use of such assays to guide de-escalation therapy requires caution.
MRD-based de-escalation strategies, whether ctDNA testing immediately after curative therapy or during surveillance, require careful comparison with current standard approaches in randomized trials of non-inferiority designs to clearly demonstrate clinical utility.
Considerations for future MRD clinical trials
First, evidence demonstrating clinical utility in adjuvant therapy (whether ctDNA testing improves clinical outcomes and increases the value of clinical decision-making compared to not using ctDNA testing) should be of the highest level and can be accomplished experimentally as follows.
-
Prospective randomized clinical trial with the primary objective of evaluating the use of ctDNA assays to guide adjuvant therapy or monitoring strategies.
-
Prospective-retrospective study using blood samples collected in prospective trials not primarily intended to assess ctDNA-guided management (usually as an exploratory endpoint). Two or more independent studies with similar results are needed to determine clinical utility.
Notably, the clinical utility of ctDNA is highly dependent on the type of disease, stage, existing treatments that are effective in eradicating MRD, and intended use. Multiple ctDNA-based randomized clinical trials are currently underway to determine the clinical utility of ctDNA in early-stage solid tumors. In general, ctDNA-guided assays can be performed in the following clinical settings.
-
Several weeks after definitive therapy: To investigate whether ctDNA testing can be used to de-escalate therapy in ctDNA-negative patients and/or intensify therapy in ctDNA-positive patients. Non-inferiority (for descending strategies) and superiority (for ascending strategies) designs can be used.
-
Shortly after completion of standard adjuvant therapy: The primary objective was to investigate whether additional “second-line” or “post-adjuvant” new therapies could improve cure rates in patients with detectable ctDNA but no imaging evidence of disease.
-
During surveillance: To determine whether ctDNA-guided surveillance can detect recurrence earlier, allow more patients to undergo radical metastatic resection, or improve patient survival compared to standard surveillance protocols.
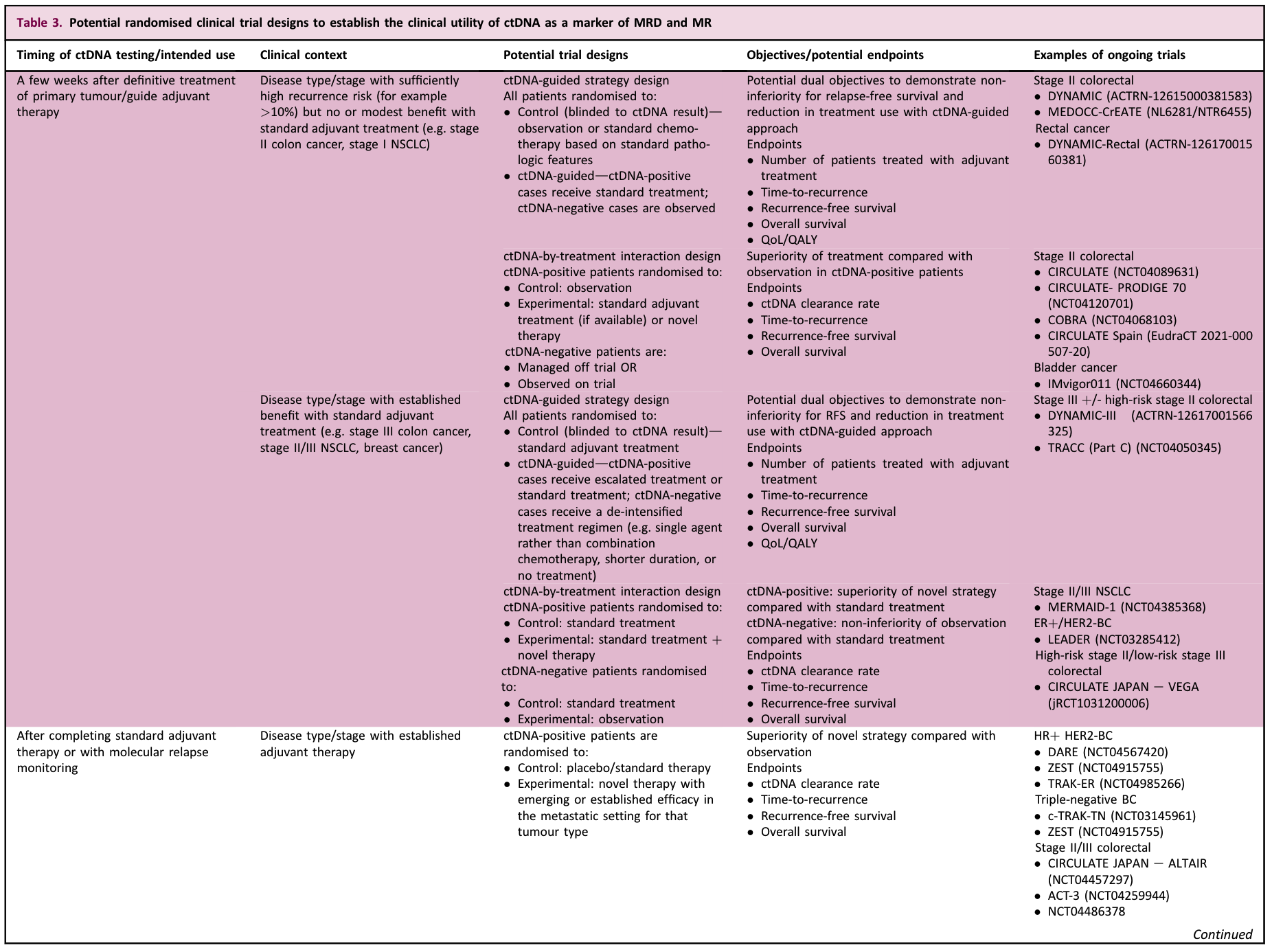
Early-stage tumor MRD/MR recommendations
-
ctDNA testing using a validated assay after curative treatment of early-stage tumors can predict the risk of recurrence, and several studies have demonstrated its clinical validity.
-
There is insufficient prospective clinical evidence that MRD-guided therapy improves efficacy or safely de-escalates therapy in routine practice.
Assist in the initial diagnosis of tumors in the early or late stage
ctDNA testing can be used as part of a diagnostic workflow for patients suspected of having tumors on imaging.
In patients with tumors that are difficult to biopsy, genotyping using ctDNA testing can be used to identify mutations and confirm the presence of the tumor. In this case, the clonal hematopoietic mutation cannot be confused with the pathogenic mutation, so the use of specialized testing methods is critical.
For patients with aggressive tumors, ctDNA testing may lead to earlier initiation of targeted therapy than tissue biopsy. For ctDNA assays used for diagnosis, the problem of false negatives must be considered.
Screen asymptomatic people
The ultimate application of ctDNA in cancer therapy may be the ability to detect early cancer and precancerous lesions in asymptomatic populations, and then take related measures to improve cure rates and even prevent the development of invasive tumors.
Achieving high specificity while maintaining clinical sensitivity is necessary for standardized screening tools to become a reality with sufficient evidence from large population-based studies.
The use of ctDNA remains technically challenging because of the low ctDNA content of early-stage tumors. Furthermore, ideally ctDNA-based screening should also provide information on the tissue of origin of the cancer, which is far from mature at this stage.
There has been a lot of work in this area, and studies have shown that high-sensitivity sequencing modalities can be combined with protein biomarkers, fragment length characterization, and methylation detection. However, ctDNA testing cannot be considered an effective use for ctDNA testing as a screening tool for various cancers until data from large studies in screening populations have confirmed it.
The above are several application scenarios of ctDNA mentioned by ESMO, and the corresponding CACA gives the following three consensuses.
At present, there is clinical evidence that ctDNA NGS detection can be applied to the companion diagnosis of advanced solid tumors such as lung cancer, breast cancer, prostate cancer, and ovarian cancer. However, the involved driver genes and their mutation types and corresponding analysis systems are strictly limited. When applying for indications, it is recommended to fully inform the patient about the necessity of testing, testing costs, and limitations. ctDNA NGS testing has been recommended by domestic and foreign experts or guidelines as an alternative to genetic testing of various advanced malignant tumor tissues, but high-level evidence-based evidence is still needed to formulate clinical treatment strategies in real time based on its analysis results. (CACA Consensus)
Quantitative and dynamic analysis of ctDNA levels based on NGS detection is expected to become an emerging approach for efficacy assessment after the initiation of molecular targeting or immune checkpoint inhibitor therapy for advanced solid tumors. ctDNA MRD detection is a new application field of personalized technology, and it is still difficult to establish a general technical standard. It is urgent to verify its clinical utility through large-sample, multi-center and prospective clinical trials. When ctDNA NGS testing is used clinically for targeted or immunotherapy evaluation and stratification, it is recommended to be fully informed about the value, limitations, and cost of testing. (CACA Consensus)
In a clinical setting, ctDNA NGS assays can be used to identify resistance mechanisms to molecularly targeted therapies, especially for patients with difficult and complex tumors, and the results can aid subsequent treatment selection decisions. The mechanisms of acquired resistance to immune checkpoint inhibitor therapy are complex, and tumor subclonal evolution under therapeutic selection pressure is only part of the reason, and ctDNA-targeted sequencing, whole-exome and/or whole-genome testing are only used as translational research tools. one. (CACA Consensus)
summary
Liquid biopsies, especially ctDNA testing, are increasingly being used in clinical practice.
There is sufficient evidence that genotyping in advanced cancer can be used to guide treatment, especially when tissue biopsy is suboptimal or timing is critical.
The use of ctDNA in the clinic must take into account the sensitivity of detection, especially fusion and copy number variations, which are less sensitive. Further development of better assays to accurately distinguish between true and false positive/negative results remains a major priority for future genotyping testing.
ctDNA assays have theoretical advantages in capturing spatial and temporal tumor heterogeneity in patients more accurately than tissue sequencing. More important clinical trials are now needed to assess how detecting this heterogeneity can provide clinically useful information to improve treatment.
Due to the lack of useful evidence, ctDNA testing is not recommended for other possible purposes such as early screening, MRD assessment, and early assessment of treatment response. New technologies currently under development, such as methylation-based, fragment-length-based sequencing, or novel ultra-sensitive mutation detection methods, have the potential to optimize these applications. Multiple ongoing clinical trials may also provide an evidence base for decision-making using ctDNA testing in a variety of clinical settings.
The above is the first part of the ctDNA learning trilogy, the sorting and learning recommended by ESMO and CACA experts. In the next article, we will study the related research of high-depth whole-genome ctDNA sequencing in prostate cancer , and look at the feasibility and application advantages of high-depth whole-genome deep ctDNA detection.
See you next time.
The author of this article : Bear thinking about problems
Copyright notice : Unless otherwise stated, all articles on this blog are licensed under the Creative Commons Attribution-Non-Commercial-No Derivatives 4.0 International License Agreement (CC BY-NC-ND 4.0) .
This article is reproduced from: https://kaopubear.top/blog/2022-08-02-ctdna-esmo-caca-recommendation/
This site is for inclusion only, and the copyright belongs to the original author.